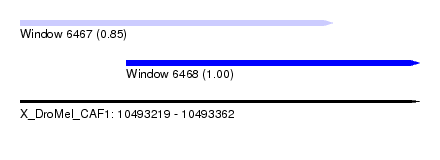
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,493,219 – 10,493,362 |
| Length | 143 |
| Max. P | 0.998263 |

| Location | 10,493,219 – 10,493,331 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10493219 112 + 22224390 CCCUCAUUUUUUCUUUUU--UUUUUUUUUUGCCACCCAACGUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUU ..................--.......((((.........(((((.......))))).(((((((((((((((.(((....).))..)))))))..)))))))).))))..... ( -21.40) >DroSec_CAF1 24437 111 + 1 CCGUCAUUUUUUCUUUUUCUUUUUUUUUUUGCCACUCAACGUGUAGUUUCUGUGCACUU---GUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUU ..(((........................(((.((..(((.....)))...))))).((---(((((((((((.(((....).))..)))))))..)))))).....))).... ( -22.10) >DroSim_CAF1 24931 107 + 1 CCGUCAUUUUUUCUUU-------UUUUUUUGCCACUCAACGUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUU ..(((...........-------......(((.((..(((.....)))...)))))..(((((((((((((((.(((....).))..)))))))..))))))))...))).... ( -24.50) >DroYak_CAF1 26243 108 + 1 CC--CAUUUUU-GUU-UU--UCGUUUUUUUGCCGCCCAACGUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUU ..--((.((((-(((-((--(.(((...((((((((.((((((((.......))))).((....))...))).)))))))).))))))))))))).))................ ( -29.60) >consensus CCGUCAUUUUUUCUUUUU__UUUUUUUUUUGCCACCCAACGUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUU ...........................((((.........(((((.......))))).(((((((((((((((.(((....).))..)))))))..)))))))).))))..... (-19.30 = -20.05 + 0.75)


| Location | 10,493,257 – 10,493,362 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
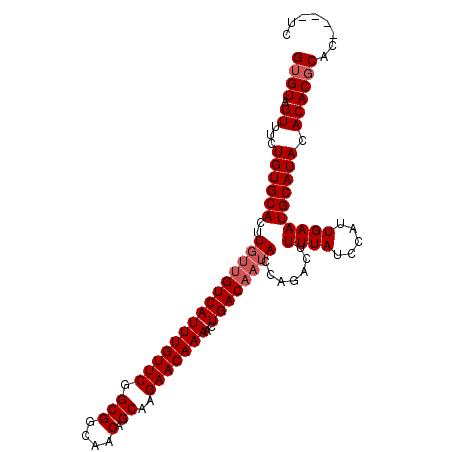
>X_DroMel_CAF1 10493257 105 + 22224390 GUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUUAUCCAUUGAAUGCAUACACACGCACUCACUC ((((.((.(.((((((..(((((((((((((((.(((....).))..)))))))..)))))))).......((((.....)))))))))).).))))))...... ( -26.60) >DroSec_CAF1 24477 98 + 1 GUGUAGUUUCUGUGCACUU---GUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUUAUCCAUUGAAUGCAUACACACGCAC----UA ((((.((.(.((((((.((---(((((((((((.(((....).))..)))))))..)))))).........((((.....)))))))))).).))))))----.. ( -24.20) >DroSim_CAF1 24964 101 + 1 GUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUUAUCCAUUGAAUGCAUACACACGCAC----UC ((((.((.(.((((((..(((((((((((((((.(((....).))..)))))))..)))))))).......((((.....)))))))))).).))))))----.. ( -26.60) >DroEre_CAF1 26066 101 + 1 GAGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUUAUCCAUUGAAUGCAUACACACGCAC----UC ((((.((.(.((((((..(((((((((((((((.(((....).))..)))))))..)))))))).......((((.....)))))))))).).))..))----)) ( -26.30) >DroYak_CAF1 26277 101 + 1 GUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUUAUCCAUUGAAUGCAUACACACGCAC----UC ((((.((.(.((((((..(((((((((((((((.(((....).))..)))))))..)))))))).......((((.....)))))))))).).))))))----.. ( -26.60) >consensus GUGUAGUUUCUGUGCACUUGUUGUCAUUUGUUCGGCGGCAACAGCAAGAACAAAACUGACAAUACCAGACUUUUAUCCAUUGAAUGCAUACACACGCAC____UC ((((.((...((((((..(((((((((((((((.(((....).))..)))))))..)))))))).......((((.....)))))))))).))))))........ (-23.74 = -24.54 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:16 2006