
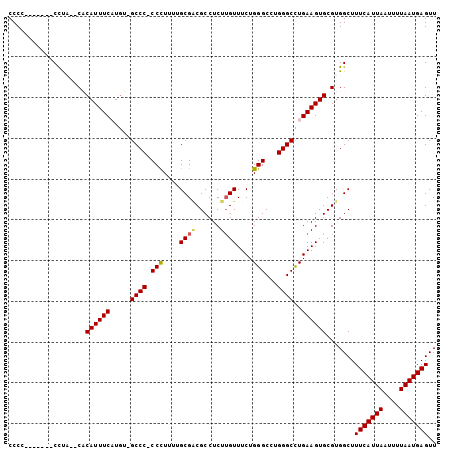
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,492,935 – 10,493,153 |
| Length | 218 |
| Max. P | 0.940191 |

| Location | 10,492,935 – 10,493,038 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -18.79 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10492935 103 + 22224390 CCCCCUCCCCCCCCUCACACAUUUCAUAU-GCCC-CCUUUUUGCGGCGCCUCUUGUUUCUGGGCCUGGGCUUGAAGUGCGUGGCUUUCAUUAAUUUUAAUGAGUU ..............((((.(((((((...-((((-.(.....).(((.((..........))))).)))).))))))).))))..(((((((....))))))).. ( -27.00) >DroSec_CAF1 24165 94 + 1 CCCC-------CCUU--CACAUUUCAUGU-GCCC-CCCUUUUGCGCCGCCUCUAGUUUCUGGGCCUGGGCCUGAAGUGCGUGGCUUUCAUUAAUUUUAAUGAGUU ....-------((..--(.(((((((.(.-((((-(((....((..........))....)))...)))))))))))).).))..(((((((....))))))).. ( -23.90) >DroEre_CAF1 25758 95 + 1 CCCA-------CUUA--CCCAUUUCUUGU-GCCCCCCCUUUUGCGACGCCUCUUGUUUCUGGGCCUGGGCCUGAAGUGCGUGGCUUUCAUUAAUUUUAAUGAGUU .(((-------(...--..((((((..(.-((((.(((....((((......))))....)))...))))).)))))).))))..(((((((....))))))).. ( -25.70) >DroYak_CAF1 25962 95 + 1 CCC--------CUUA--CCCAUUUCUUGGGGCCCCCCCUUUGGCGACGCCUCUUGUUUCUGGGCCUGGGCCUGAAGUGCGUGGCUUUCAUUAAUUUUAAUGAGUU ...--------....--.((((..(((.((((((.(((...(((((......)))))...)))...)))))).)))...))))..(((((((....))))))).. ( -30.50) >consensus CCCC_______CCUA__CACAUUUCAUGU_GCCC_CCCUUUUGCGACGCCUCUUGUUUCUGGGCCUGGGCCUGAAGUGCGUGGCUUUCAUUAAUUUUAAUGAGUU ...................((((((.....((((.(((....((((......))))....)))...))))..)))))).......(((((((....))))))).. (-18.79 = -18.97 + 0.19)


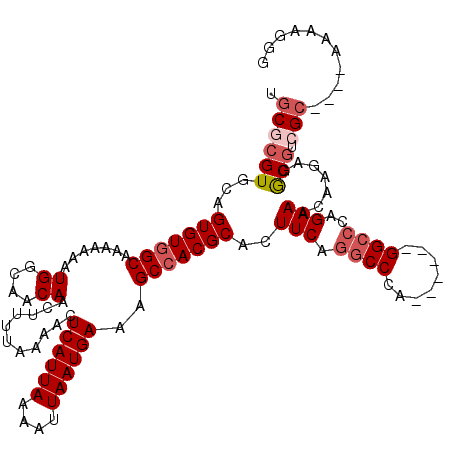
| Location | 10,492,968 – 10,493,078 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -25.02 |
| Energy contribution | -26.05 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10492968 110 - 22224390 UGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAAGCCCA------GGCCCAGAAACAAGAGGCGCCGC----AAAAAGG ((((.((((.(((((((.......((....))...........((((((....))))))..)))))))..(((..((...------.))...))).......)))))))----)...... ( -33.20) >DroSec_CAF1 24189 110 - 1 UGCACGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA------GGCCCAGAAACUAGAGGCGGCGC----AAAAGGG .....((((.(((((((.......((....))...........((((((....))))))..)))))))..(((.(((...------.)))..)))..........))))----....... ( -31.00) >DroSim_CAF1 24677 116 - 1 UGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCAGGCCCAGGCCCAGAAACAAGAGGCGGCGC----AAAAGGG (((((.(((.(((((((.......((....))...........((((((....))))))..)))))))..(((.((((........))))..))).......)))))))----)...... ( -37.80) >DroEre_CAF1 25783 110 - 1 UGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA------GGCCCAGAAACAAGAGGCGUCGC----AAAAGGG (((((((...(((((((.......((....))...........((((((....))))))..)))))))..(((.(((...------.)))..))).......))).)))----)...... ( -33.30) >DroYak_CAF1 25987 110 - 1 UGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA------GGCCCAGAAACAAGAGGCGUCGC----CAAAGGG ((.((((((.(((((((.......((....))...........((((((....))))))..)))))))..(((.(((...------.)))..))).......))).)))----))..... ( -32.90) >DroAna_CAF1 26559 113 - 1 GGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCGACGCACUUCAGGCCCA------GGACCAGAAACAAGAGACCUAGACCGAAAAAA-A ((.((.((.((((((((.......((....))...........((((((....))))))..))..)))))).)).))))(------((.((........).).)))............-. ( -23.50) >consensus UGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA______GGCCCAGAAACAAGAGGCGUCGC____AAAAGGG .((((((...(((((((.......((....))...........((((((....))))))..)))))))..(((.((((........))))..))).......))).)))........... (-25.02 = -26.05 + 1.03)


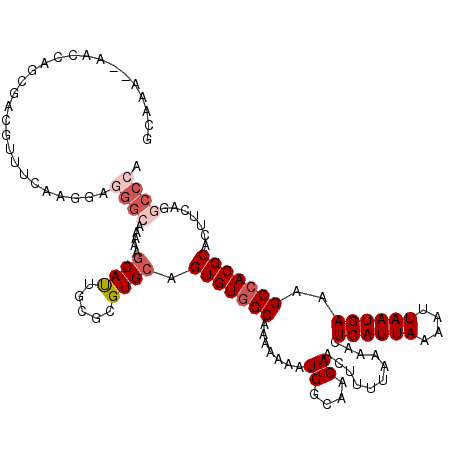
| Location | 10,492,998 – 10,493,118 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -24.37 |
| Energy contribution | -25.57 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10492998 120 - 22224390 GCAAAAAAAACAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAAGCCCA ((..........)).............((((.....((((.....)))).(((((((.......((....))...........((((((....))))))..))))))).......)))). ( -31.60) >DroSec_CAF1 24219 118 - 1 GCAAA--AAACAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCACGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA ((...--.....)).............((((.....((((.....)))).(((((((.......((....))...........((((((....))))))..))))))).......)))). ( -32.30) >DroSim_CAF1 24713 118 - 1 GCAAA--AACCAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA ((...--.....)).............((((.....((((.....)))).(((((((.......((....))...........((((((....))))))..))))))).......)))). ( -32.30) >DroEre_CAF1 25813 118 - 1 GCAAA--AACCAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA ((...--.....)).............((((.....((((.....)))).(((((((.......((....))...........((((((....))))))..))))))).......)))). ( -32.30) >DroYak_CAF1 26017 106 - 1 GCAAA--AACCAGCGACGUUUCAA------------GCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA .....--.....((((.((.....------------)).))))((.((..(((((((.......((....))...........((((((....))))))..)))))))....)).))... ( -26.10) >DroAna_CAF1 26592 102 - 1 ---AA--AACCAA---------AAGGGUGG----AAACACGGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCGACGCACUUCAGGCCCA ---..--..((..---------..))(((.----...)))((.((.((.((((((((.......((....))...........((((((....))))))..))..)))))).)).)))). ( -30.10) >consensus GCAAA__AACCAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAAAACUCAUUAAAAUUAAUGAAAGCCACGCACUUCAGGCCCA ...........................((((.....((((.....)))).(((((((.......((....))...........((((((....))))))..))))))).......)))). (-24.37 = -25.57 + 1.20)


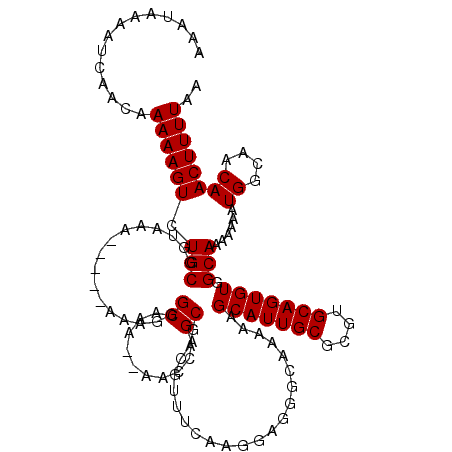
| Location | 10,493,038 – 10,493,153 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10493038 115 - 22224390 AAAUAAAAUCAACAAAAAGUCUGCGGUAAA-----AAAGGGCAAAAAAAACAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAA ..................(((.((.((...-----..............)).))))).......(((((((.....(((((((....))))))).)).......((....)).))))).. ( -18.13) >DroSec_CAF1 24259 113 - 1 AAAUAAAAUCAACAAAAAGUCUGCGGUAAA-----AAAGGGCAAA--AAACAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCACGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAA ..............((((((.(((.((...-----.....((...--.....))...............((.....(((((((....))))))).))......)).)))...)))))).. ( -19.10) >DroSim_CAF1 24753 113 - 1 AAAUAAAAUCAACAAAAAGUCUGCGGUAAA-----AAAGGGCAAA--AACCAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAA ..................(((.(((((...-----..........--.))).))))).......(((((((.....(((((((....))))))).)).......((....)).))))).. ( -20.64) >DroEre_CAF1 25853 113 - 1 AAAUAAAAUCAACAAAAAGUCUGCGGCAAA-----AAAGGGCAAA--AACCAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAA ..............((((((.(((.(((..-----...((.....--..)).((.((((..(((.....((.....)).))).))))))..))).)))......((....)))))))).. ( -21.40) >DroYak_CAF1 26057 106 - 1 AAAUAAAAUCAACAAAAAGUCUGCGGCAAAAAAAAAAAGGGCAAA--AACCAGCGACGUUUCAA------------GCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAA ..............(((((((((((((...........((.....--..)).((((.((.....------------)).)))))).)))))((((.((......)).).))))))))).. ( -19.90) >consensus AAAUAAAAUCAACAAAAAGUCUGCGGUAAA_____AAAGGGCAAA__AACCAGCGACGUUUCAAGGAGGGCAAAAAGCAUUGCGCGUGCAGUGUGGCAAAAAAAUGGCAACAACUUUUAA ..............((((((.(((................((..........))......................(((((((....))))))).)))......((....)))))))).. (-16.08 = -16.08 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:14 2006