
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,491,703 – 10,491,819 |
| Length | 116 |
| Max. P | 0.950896 |

| Location | 10,491,703 – 10,491,795 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.60 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.08 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10491703 92 - 22224390 AAUACACAAUUUUGUAG-----CACACACGCCACGUU-UGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUACAAG------UACAACCAAAAAUCA ..........(((((((-----((.((((((((....-))))).)))..)))..(((.......)))..........))))))------............... ( -18.30) >DroSec_CAF1 22957 92 - 1 AAUACACAAUUUUGUAG-----CACACACGCCACGUU-UGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUACAAG------UAAAACCAAAAAUCA ..........(((((((-----((.((((((((....-))))).)))..)))..(((.......)))..........))))))------............... ( -18.30) >DroSim_CAF1 23138 92 - 1 AAUACACAAUUUUGUAG-----CACACACGCCACGUU-UGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUACAAG------UAAAACCAAAAAUCA ..........(((((((-----((.((((((((....-))))).)))..)))..(((.......)))..........))))))------............... ( -18.30) >DroEre_CAF1 24505 97 - 1 ACUACCCA-UUUUGUAG-----CACACACGCCACGUU-UGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGGACAAGUACAAGUACAACCAAAAAUCA .....((.-...(((((-----.(((.((((((....-))))))))).))))).(((.......))).........))....(((....)))............ ( -20.20) >DroYak_CAF1 24658 98 - 1 AAUACACAGCUUUGUGGCACGGCACACACGCCACGUUUUGGCGUUGUCUUGCACGUGGCCCCAACACAAGGAACAAGGACAAG------UACAACCAAAAAUCA ..(((....(((((((((((((((.((((((((.....))))).)))..))).))).)))((.......)).))))))....)------))............. ( -26.90) >consensus AAUACACAAUUUUGUAG_____CACACACGCCACGUU_UGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUACAAG______UACAACCAAAAAUCA .........((((((.(.....)......(((((((....(((......))))))))))......))))))................................. (-14.40 = -14.08 + -0.32)

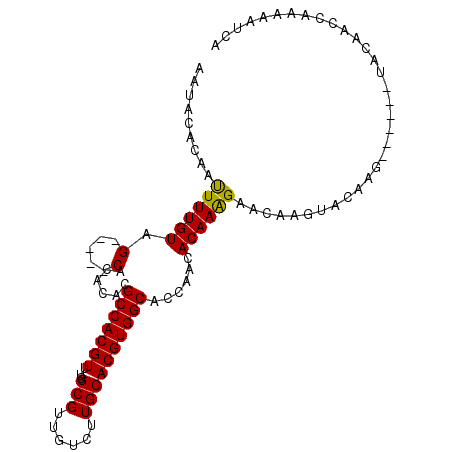
| Location | 10,491,721 – 10,491,819 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.57 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -20.52 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10491721 98 - 22224390 UAUGG--GCCCAAUAACACACA------------UACAAAUACACAAUUUUGUAGCACACACGCCACGUUUGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUAC ..(((--((((...........------------((((((........))))))(((.((((((((....))))).)))..)))..).))).)))................. ( -24.80) >DroSec_CAF1 22975 98 - 1 UAGGG--GCCCAAUCACACACA------------UACAAAUACACAAUUUUGUAGCACACACGCCACGUUUGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUAC ..((.--((((...........------------((((((........))))))(((.((((((((....))))).)))..)))..).))).)).................. ( -24.10) >DroSim_CAF1 23156 98 - 1 UAGGG--GCCCAAUCACACGCA------------UACAAAUACACAAUUUUGUAGCACACACGCCACGUUUGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUAC ..((.--((((........((.------------((((((........))))))))(((.((((((....))))))))).......).))).)).................. ( -24.30) >DroEre_CAF1 24529 111 - 1 CAGGGUGGCCCAAUCACACAACUACAAAUACAAAUACAACUACCCA-UUUUGUAGCACACACGCCACGUUUGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGGAC ..((((((...............................)))))).-((((((.(((.((((((((....))))).)))..)))..(((.......))).....)))))).. ( -24.98) >consensus UAGGG__GCCCAAUCACACACA____________UACAAAUACACAAUUUUGUAGCACACACGCCACGUUUGGCGUUGUCUUGCACGUGGCACCAACACAAAGAACAAGUAC ..((...((((.......................((((((........))))))(((.((((((((....))))).)))..)))..).))).)).................. (-20.52 = -21.02 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:10 2006