
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,491,325 – 10,491,433 |
| Length | 108 |
| Max. P | 0.866952 |

| Location | 10,491,325 – 10,491,433 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -31.46 |
| Energy contribution | -32.26 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10491325 108 + 22224390 CGCUACCCCUCCCCC-----CAGCC-CCCGACACUGCACACGCUGUAAUUGUGGCCAGCUUUGAGCUGAGAAUUUUCCCUCAGCUUGUGUGCGUGUUGGCGAGAUUGCUGUCAA ...............-----(((((-((((((((.((((((((((..........))))...((((((((........))))))))))))))))))))).).)...)))).... ( -39.90) >DroSec_CAF1 22582 114 + 1 CGCUACCCCUCCACCACCCCCUCCGCCCAAGCACUGCACACGCUGUAAUUGUGGCCAGCUUUGAGCUGAGAAUUUUCCCUGAGCUUGUGUGCGUGCUUGCGAGAUUGCUGUCAA .......................(((..((((((.((((((((((..........))))...(((((.((........)).)))))))))))))))))))).(((....))).. ( -34.50) >DroSim_CAF1 22768 114 + 1 CGCUACCCCUCCGCCACCCCCUCCGCCCAAGCACUGCACACGCUGUAAUUGUGGCCAGCUUUGAGCUGAGAAUUUUCCCUGAGCUUGUGUGCGUGCUUGCGAGAUUGCUGUCAA .((.....(((.((..........)).(((((((.((((((((((..........))))...(((((.((........)).)))))))))))))))))).)))...))...... ( -34.70) >DroEre_CAF1 24185 94 + 1 CGCUCCCCCU--------------------GCACUGCACACGCUGUAAUUGUGGCCAGCUUUGAGCUGAGAAUUUCCCCUCAGCUUGUGUGCGUGUUUGCGAGAUUGCUGUCAA (((.......--------------------((((.((((((((((..........))))...((((((((........))))))))))))))))))..))).(((....))).. ( -33.50) >DroYak_CAF1 24224 94 + 1 CGCUCCACCU--------------------GCACUGCACACGCUGUAAUUGUGGCCAGCUUUGAGCUGAGACUUACCCUUCAGCUUGUGUGCGUGUUAGCGAGAUUGCUGUCAA ((((......--------------------((((.((((((((((..........))))...((((((((........)))))))))))))))))).)))).(((....))).. ( -31.90) >consensus CGCUACCCCUCC_CC_____C__C__CC__GCACUGCACACGCUGUAAUUGUGGCCAGCUUUGAGCUGAGAAUUUUCCCUCAGCUUGUGUGCGUGUUUGCGAGAUUGCUGUCAA ...........................(((((((.((((((((((..........))))...((((((((........)))))))))))))))))))))...(((....))).. (-31.46 = -32.26 + 0.80)

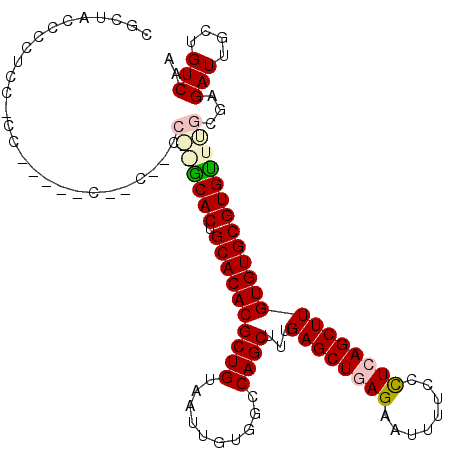

| Location | 10,491,325 – 10,491,433 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -29.33 |
| Energy contribution | -30.77 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10491325 108 - 22224390 UUGACAGCAAUCUCGCCAACACGCACACAAGCUGAGGGAAAAUUCUCAGCUCAAAGCUGGCCACAAUUACAGCGUGUGCAGUGUCGGG-GGCUG-----GGGGGAGGGGUAGCG ......((.(((((.((.((((((((((.(((((((((....)))))))))....((((..........)))))))))).))))(((.-..)))-----...)).))))).)). ( -40.50) >DroSec_CAF1 22582 114 - 1 UUGACAGCAAUCUCGCAAGCACGCACACAAGCUCAGGGAAAAUUCUCAGCUCAAAGCUGGCCACAAUUACAGCGUGUGCAGUGCUUGGGCGGAGGGGGUGGUGGAGGGGUAGCG ((.((.((..((((((.(((((((((((.((((..((((....))))))))....((((..........)))))))))).)))))...)).))))..)).)).))......... ( -39.60) >DroSim_CAF1 22768 114 - 1 UUGACAGCAAUCUCGCAAGCACGCACACAAGCUCAGGGAAAAUUCUCAGCUCAAAGCUGGCCACAAUUACAGCGUGUGCAGUGCUUGGGCGGAGGGGGUGGCGGAGGGGUAGCG ......((.(((((.(((((((((((((.((((..((((....))))))))....((((..........)))))))))).))))))).((.(......).))...))))).)). ( -41.60) >DroEre_CAF1 24185 94 - 1 UUGACAGCAAUCUCGCAAACACGCACACAAGCUGAGGGGAAAUUCUCAGCUCAAAGCUGGCCACAAUUACAGCGUGUGCAGUGC--------------------AGGGGGAGCG ......((..(((((((.....((((((.(((((((((....)))))))))....((((..........))))))))))..)))--------------------.))))..)). ( -36.60) >DroYak_CAF1 24224 94 - 1 UUGACAGCAAUCUCGCUAACACGCACACAAGCUGAAGGGUAAGUCUCAGCUCAAAGCUGGCCACAAUUACAGCGUGUGCAGUGC--------------------AGGUGGAGCG ......((..((.((((..(((((((((.((((((.((.....))))))))....((((..........)))))))))).))).--------------------.)))))))). ( -32.20) >consensus UUGACAGCAAUCUCGCAAACACGCACACAAGCUGAGGGAAAAUUCUCAGCUCAAAGCUGGCCACAAUUACAGCGUGUGCAGUGC__GG__G__G_____GG_GGAGGGGUAGCG ......((.(((((....((((((((((.((((((((......))))))))....((((..........)))))))))).)))).....................))))).)). (-29.33 = -30.77 + 1.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:08 2006