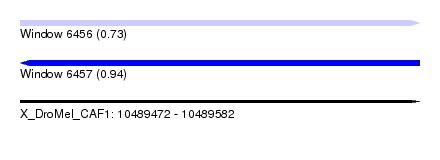
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,489,472 – 10,489,582 |
| Length | 110 |
| Max. P | 0.935021 |

| Location | 10,489,472 – 10,489,582 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.64 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10489472 110 + 22224390 AAUAUCCUGAGCAACAUAUGUGAACU-UUGGUCUUUUCCCAUUUUCAUGUUUGCUGCUUACUUAU------AUGUGCAUCCACUUGUUUGUCUGACCAUUUUCUAUGGCCAUGAUCU .((((..((((((.((.(((((((..-.(((.......)))..))))))).)).))))))...))------))(((....)))......(((((.((((.....)))).)).))).. ( -21.00) >DroSec_CAF1 20201 110 + 1 AAUAACCAGAGCAACAAAUGUGAACU-UUGGUCUUUUCCUAUUUUCAUGUUUGCUGCUUACUUAU------AUGUGCAUCCACUUGUUUGUCUGACCAUUUUCUAUGGCCAUGAUCU ........(((((.((((((((((..-..((......))....)))))))))).)))))((..((------(.(((....))).)))..)).((.((((.....)))).))...... ( -23.20) >DroSim_CAF1 20830 110 + 1 AAUAUCCAGAGCAACAAAUGUGAACU-UUGGUCUUUUCCUAUUUUCAUGUUUGCUGCUUACUUAU------AUGUGCAUCCACUUGUUUGUCUGACCAUUUUCUAUGGCCAUGAUCU ........(((((.((((((((((..-..((......))....)))))))))).)))))((..((------(.(((....))).)))..)).((.((((.....)))).))...... ( -23.20) >DroEre_CAF1 21540 109 + 1 AA-AUCCUGUGCAACAUAUGUGAACU-UUGGUCUUUUCCUAUUUUUAUGUUUGCUGCUUACUUAU------AUGUGCAUCCACUUGUUUGUCUGACCAUUUUCUAUGGCCAUGAUCU ..-.......(((.((.(((((((..-..((......))....))))))).)).)))..((..((------(.(((....))).)))..)).((.((((.....)))).))...... ( -15.00) >DroYak_CAF1 22530 116 + 1 AAUAUCCUGAACAACAUAUGUGAAUUUUUGGUCUUUUCCUAUUUUCACAU-UGCUGCUUACUUAUAGUCGUAUGUUCAUCCAUUUGUUUGUCUGACCAUUUGCCAUGGCCAUGAUCU .......((((((....(((((((.....((......))....)))))))-(((.(((.......))).)))))))))...........(((((.((((.....)))).)).))).. ( -20.50) >consensus AAUAUCCUGAGCAACAUAUGUGAACU_UUGGUCUUUUCCUAUUUUCAUGUUUGCUGCUUACUUAU______AUGUGCAUCCACUUGUUUGUCUGACCAUUUUCUAUGGCCAUGAUCU ........(((((.((.(((((((.....((......))....))))))).)).)))))..............................(((((.((((.....)))).)).))).. (-16.72 = -16.64 + -0.08)


| Location | 10,489,472 – 10,489,582 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10489472 110 - 22224390 AGAUCAUGGCCAUAGAAAAUGGUCAGACAAACAAGUGGAUGCACAU------AUAAGUAAGCAGCAAACAUGAAAAUGGGAAAAGACCAA-AGUUCACAUAUGUUGCUCAGGAUAUU ..(((.(((((((.....))))))).........(((....)))..------.......(((((((....((((..(((.......))).-..))))....)))))))...)))... ( -24.60) >DroSec_CAF1 20201 110 - 1 AGAUCAUGGCCAUAGAAAAUGGUCAGACAAACAAGUGGAUGCACAU------AUAAGUAAGCAGCAAACAUGAAAAUAGGAAAAGACCAA-AGUUCACAUUUGUUGCUCUGGUUAUU ...((.(((((((.....))))))))).......(((....)))..------((((.(((((((((((..((((....((......))..-..))))..))))))))).)).)))). ( -26.90) >DroSim_CAF1 20830 110 - 1 AGAUCAUGGCCAUAGAAAAUGGUCAGACAAACAAGUGGAUGCACAU------AUAAGUAAGCAGCAAACAUGAAAAUAGGAAAAGACCAA-AGUUCACAUUUGUUGCUCUGGAUAUU ..(((.(((((((.....)))))))......((.(((....)))..------.......(((((((((..((((....((......))..-..))))..))))))))).)))))... ( -25.70) >DroEre_CAF1 21540 109 - 1 AGAUCAUGGCCAUAGAAAAUGGUCAGACAAACAAGUGGAUGCACAU------AUAAGUAAGCAGCAAACAUAAAAAUAGGAAAAGACCAA-AGUUCACAUAUGUUGCACAGGAU-UU (((((.(((((((.....))))))).........(((....)))..------........((((((............((......))..-.(....)...))))))....)))-)) ( -19.40) >DroYak_CAF1 22530 116 - 1 AGAUCAUGGCCAUGGCAAAUGGUCAGACAAACAAAUGGAUGAACAUACGACUAUAAGUAAGCAGCA-AUGUGAAAAUAGGAAAAGACCAAAAAUUCACAUAUGUUGUUCAGGAUAUU ..(((.(((((((.....)))))))..............(((((.(((........)))...((((-(((((((....((......)).....))))))).))))))))).)))... ( -25.20) >consensus AGAUCAUGGCCAUAGAAAAUGGUCAGACAAACAAGUGGAUGCACAU______AUAAGUAAGCAGCAAACAUGAAAAUAGGAAAAGACCAA_AGUUCACAUAUGUUGCUCAGGAUAUU ...((.(((((((.....))))))))).......(((......)))..............((((((....((((....((......)).....))))....)))))).......... (-18.34 = -18.22 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:06 2006