
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,483,933 – 10,484,205 |
| Length | 272 |
| Max. P | 0.976619 |

| Location | 10,483,933 – 10,484,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10483933 120 + 22224390 GAAUGUUUCGCCUACAUUUUAUUACCUCCAUUCAUUGUCAUGGUCGGCCCUCUUUUCUGGCAUUUAGAUGCAUUCGGUUACGCUUCCUUUGGAGGCACUUCCUCAUCACGCCAUUCCAUC ((((((.((...............(..((((........))))..)(((.........))).....)).))))))((....((((((...))))))....)).................. ( -23.70) >DroSec_CAF1 14695 120 + 1 GAAUGUUUCGCGUACAUUUUAUUACCUCCAUUCAUUGUCAUGGUCGGCCCUCUUUUCUGGCAUUUAGCUGCAUUCGGUAACGCUUCCUUUGGAGGCACUUACUCAUCGCGCCAUUCCAUC (((((...((((.........(((((.((((........))))...(((.........)))..............))))).((((((...))))))..........)))).))))).... ( -26.40) >DroSim_CAF1 14888 120 + 1 GAAUGUUUCGCCUACAUUUUAUUACCUCCAUUCAUUGUCAUGGUCGGCCCUCUUUUCUGGCAUUUAGCUGCAUUCGGUAACGCUUCCUUUGGAGGCACUUCCUCAUCGCGCCAUUCCAUC (((((...(((..........(((((.((((........))))...(((.........)))..............))))).((((((...))))))...........))).))))).... ( -24.40) >DroEre_CAF1 16169 120 + 1 GAAUGUUUUGCCCCCACUUUAUUACCUCCAUUCAUUGUCAUGGUCGGCCUUCUUUACUGGCAUUUAGCUGCAUACGGUUACGCUUCCUUUGGAGGCACUUCCUCAUCACGCCGUUCCAUC (((((.......................)))))......((((.((((................((((((....)))))).((((((...)))))).............))))..)))). ( -24.30) >DroYak_CAF1 16857 120 + 1 GAAUGUUUCUCCUUCAUUUUAUUACCUCCAUUCAUUAUCCUGGUCGGCCUUCUUUUCUGGCAUUUAGCGGCAUUCGGUUACGCUUCCCUUGGAGGCACUUCCUCAUCACUCCAUUCCAUC ((((((..((..............(..(((..........)))..)(((.........)))....))..))))))((....((((((...))))))....)).................. ( -22.30) >consensus GAAUGUUUCGCCUACAUUUUAUUACCUCCAUUCAUUGUCAUGGUCGGCCCUCUUUUCUGGCAUUUAGCUGCAUUCGGUUACGCUUCCUUUGGAGGCACUUCCUCAUCACGCCAUUCCAUC ((((((..(...............(..((((........))))..)(((.........))).....)..))))))((....((((((...))))))....)).................. (-19.74 = -20.18 + 0.44)

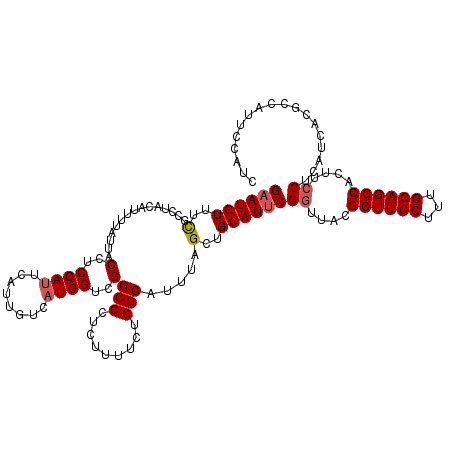
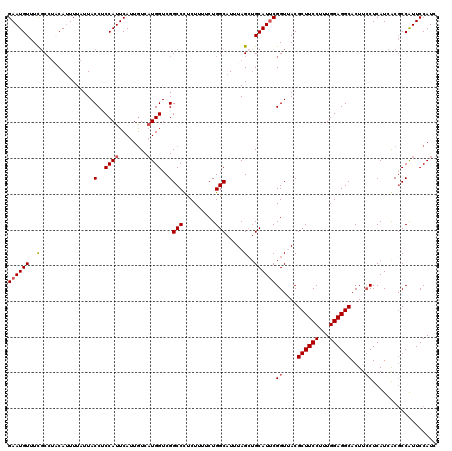
| Location | 10,484,013 – 10,484,128 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.01 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10484013 115 + 22224390 CGCUUCCUUUGGAGGCACUUCCUCAUCACGCCAUUCCAUCACCA-GCCAAGUUGCCACUUGGCC----AUAGACUCGGCUUUGCCUCUGACAUUGUGGCUUUUAAAUUGCUUAUUUGGCU .((((((...)))))).............(((((....(((...-(((((((....))))))).----........(((...)))..)))....))))).........(((.....))). ( -30.70) >DroSec_CAF1 14775 116 + 1 CGCUUCCUUUGGAGGCACUUACUCAUCGCGCCAUUCCAUCAUCAGGCCAAGUUGCCACUUGGCC----ACAGACUCGGCUUUGCCUCUGACAUUGUGGCUUUUAAAUUGCUUAUUUGGCU .((((((...)))))).............(((((..........((((((((....))))))))----.((((...(((...))))))).....))))).........(((.....))). ( -36.10) >DroSim_CAF1 14968 116 + 1 CGCUUCCUUUGGAGGCACUUCCUCAUCGCGCCAUUCCAUCAUCAGGCCAAGUUGCCACUUGGCC----ACAGACUCGGCUUUGCCUCUGACAUUGUGGCUUUUAAAUUGCUUAUUUGGCU .((((((...)))))).............(((((..........((((((((....))))))))----.((((...(((...))))))).....))))).........(((.....))). ( -36.10) >DroEre_CAF1 16249 115 + 1 CGCUUCCUUUGGAGGCACUUCCUCAUCACGCCGUUCCAUCACCA-GCCAAGUUGCCACUUGGCC----GCAGACUCGUCUUUGCCUCUGACAUUGUGGCUUUUAAAUUGCUUAUUUGGCU .((((((...))))))...........................(-(((((((.((.....((((----((((....(((.........))).))))))))........))..)))))))) ( -30.42) >DroYak_CAF1 16937 118 + 1 CGCUUCCCUUGGAGGCACUUCCUCAUCACUCCAUUCCAUCGCCA-GCCAAGUUGUCUG-UGGCCAAGUACAGACUCGGCUUUGCCUCUGACAUUGUGGCUUUUAAAUUGCUUAUUUGGCU .((((((...))))))........................((((-((((...((((.(-.(((.((((.........)))).))).).))))...)))))...((((.....))))))). ( -33.00) >consensus CGCUUCCUUUGGAGGCACUUCCUCAUCACGCCAUUCCAUCACCA_GCCAAGUUGCCACUUGGCC____ACAGACUCGGCUUUGCCUCUGACAUUGUGGCUUUUAAAUUGCUUAUUUGGCU .((((((...)))))).............(((((...........((((((......))))))......((((...(((...))))))).....))))).........(((.....))). (-26.46 = -26.94 + 0.48)

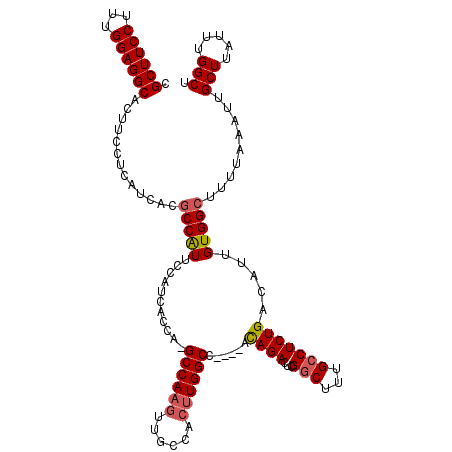

| Location | 10,484,088 – 10,484,205 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10484088 117 - 22224390 GGUUAUUAAACUGCUCGGCACGUUGCGGUUAGCCAACAGAACAGCGCUGACAACAACGACGACGACGACGAGGACUAAGCCAAAUAAGCAAUUUAAAAGCCACAAUGUCAGAGGCAA ((((.(((((.((((.(((.(((((..(((((((.........).))))))..))))).((....))...........))).....)))).))))).))))....(((.....))). ( -30.90) >DroSec_CAF1 14851 117 - 1 GGUUAUUAAACUGCUCGGCACGUUGCGGUUAGCCAACAGAACAGCGCUGACAACAACGACGACGACGACGAGGACUAAGCCAAAUAAGCAAUUUAAAAGCCACAAUGUCAGAGGCAA ((((.(((((.((((.(((.(((((..(((((((.........).))))))..))))).((....))...........))).....)))).))))).))))....(((.....))). ( -30.90) >DroEre_CAF1 16324 114 - 1 GGUUAUUAAACUGCUCGGCACGUUGCGGUUAGCCAACAGAACGGCGCUGACAAGAACGACG---ACGACGAGGACUAAGCCAAAUAAGCAAUUUAAAAGCCACAAUGUCAGAGGCAA ((((.(((((.((((((...(((((((....(((........))).((....))..)).))---))).)))((......))......))).))))).))))....(((.....))). ( -27.20) >DroYak_CAF1 17015 114 - 1 GGUUAUUAAAAUGCUCGGCGUGUUGCGGUUAGCCAACAGAACAGCGCUGACAACAAAGACG---ACGACGGGUACUAAGCCAAAUAAGCAAUUUAAAAGCCACAAUGUCAGAGGCAA ((((.(((((.((((((.(((.(((..(((((((.........).))))))..)))..)))---.)))..(((.....)))......))).))))).))))....(((.....))). ( -29.80) >consensus GGUUAUUAAACUGCUCGGCACGUUGCGGUUAGCCAACAGAACAGCGCUGACAACAACGACG___ACGACGAGGACUAAGCCAAAUAAGCAAUUUAAAAGCCACAAUGUCAGAGGCAA ((((.(((((.((((.(((.(((((..(((((((.........).))))))..))))).((....))...........))).....)))).))))).))))....(((.....))). (-27.04 = -27.35 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:01 2006