
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,480,026 – 10,480,194 |
| Length | 168 |
| Max. P | 0.999666 |

| Location | 10,480,026 – 10,480,118 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.61 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10480026 92 + 22224390 UAAAGCUGGAGCAAUAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACA ....((((.(((((.......))))).))))......(((.((((..(((((((((.(......).).))))))))...)))).)))..... ( -31.00) >DroSec_CAF1 10871 92 + 1 UAAAGCUGGAGCAAUAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAGAACA ....((((.(((((.......))))).))))......(((.((((..(((((((((.(......).).))))))))...)))).)))..... ( -31.00) >DroSim_CAF1 11085 92 + 1 UAAAGCUGGAGCAAUAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACA ....((((.(((((.......))))).))))......(((.((((..(((((((((.(......).).))))))))...)))).)))..... ( -31.00) >DroEre_CAF1 12156 92 + 1 UGAAGCUGGAGCAAUACAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACA .((.((..((((((.......)))))..(((((....((((.((((...)))).))))))))))..))..)).((((....))))....... ( -31.10) >DroYak_CAF1 12704 92 + 1 UGAAGCUGGAGCAAUAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGACGCUAUGGCGCAAAAACA ....((((.(((((.......))))).))))....((((((.((((...)))).))))))(((.(((((.....))))).)))......... ( -30.40) >consensus UAAAGCUGGAGCAAUAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACA ....((((.(((((.......))))).))))......(((.((((..(((((((((.(......).).))))))))...)))).)))..... (-29.70 = -29.90 + 0.20)



| Location | 10,480,026 – 10,480,118 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.61 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -22.12 |
| Energy contribution | -21.96 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.10 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10480026 92 - 22224390 UGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCCCAGCAUCUAGCCCAAUAUUUGGCCACAGCAAACAUUUAUUGCUCCAGCUUUA .......((((....)))).((((.....(((((..(((..........))).......)))))..(((((.......)))))...)))).. ( -22.30) >DroSec_CAF1 10871 92 - 1 UGUUCUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCCCAGCAUCUAGCCCAAUAUUUGGCCACAGCAAACAUUUAUUGCUCCAGCUUUA (((((..((((....)))).)))))....((((...)))).(((.....(((........)))...(((((.......)))))...)))... ( -23.50) >DroSim_CAF1 11085 92 - 1 UGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCCCAGCAUCUAGCCCAAUAUUUGGCCACAGCAAACAUUUAUUGCUCCAGCUUUA .......((((....)))).((((.....(((((..(((..........))).......)))))..(((((.......)))))...)))).. ( -22.30) >DroEre_CAF1 12156 92 - 1 UGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCCCAGCAUCUAGCCCAAUAUUUGGCCACAGCAAACAUGUAUUGCUCCAGCUUCA .......((((....)))).((((.....(((((..(((..........))).......)))))..(((((.......)))))...)))).. ( -22.30) >DroYak_CAF1 12704 92 - 1 UGUUUUUGCGCCAUAGCGUCGAGCACCAAGGCCAUCGGCCCAGCAUCUAGCCCAAUAUUUGGCCACAGCAAACAUUUAUUGCUCCAGCUUCA .......((((....)))).((((.....(((((..(((..........))).......)))))..(((((.......)))))...)))).. ( -20.60) >consensus UGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCCCAGCAUCUAGCCCAAUAUUUGGCCACAGCAAACAUUUAUUGCUCCAGCUUUA .......((((....)))).((((.....(((((..(((..........))).......)))))..(((((.......)))))...)))).. (-22.12 = -21.96 + -0.16)

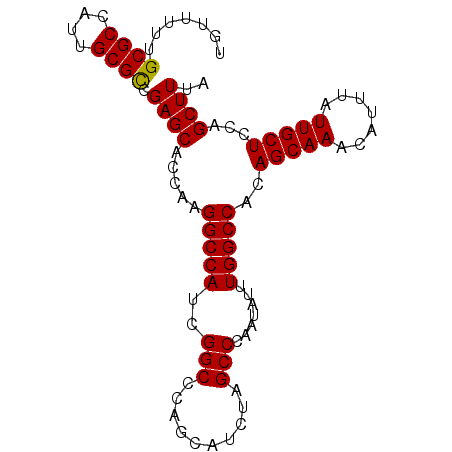
| Location | 10,480,040 – 10,480,154 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 97.37 |
| Mean single sequence MFE | -39.96 |
| Consensus MFE | -39.24 |
| Energy contribution | -39.04 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10480040 114 + 22224390 UAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACAACAGCGAACGAAAUGGGACAUCCUGUGCUCCGGAAG .....((((((((((((((....((((.((((...)))).)))))))))...((.....((((....))))....)).)))))))))((.(((((....)))))..))...... ( -41.40) >DroSec_CAF1 10885 114 + 1 UAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAGAACAACAGUGGACGAAAUGGGACAUCCUGUGCUCCGGAAG .....(((..(((((((((....((((.((((...)))).)))))))))...((.((..((((....)))).)).)).))))..)))((.(((((....)))))..))...... ( -39.60) >DroSim_CAF1 11099 114 + 1 UAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACAACAGUGGACGAAAUGGGACAUCCUGUGCUCCGGAAG .....(((..(((((((((....((((.((((...)))).)))))))))...((.....((((....))))....)).))))..)))((.(((((....)))))..))...... ( -39.60) >DroEre_CAF1 12170 114 + 1 UACAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACAACAGCGGACGAAAUGGGACAUCCUGUGCUCCGGAAA .....((((((((((((((....((((.((((...)))).)))))))))...((.....((((....))))....)).)))))))))((.(((((....)))))..))...... ( -41.00) >DroYak_CAF1 12718 114 + 1 UAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGACGCUAUGGCGCAAAAACAACAGCGGACGAAAUGGGACAUCCUGUGCUCCGGAAG .....((((((((((((((....((((.((((...)))).)))))))))...(((((.........))))).......)))))))))((.(((((....)))))..))...... ( -38.20) >consensus UAAAUGUUUGCUGUGGCCAAAUAUUGGGCUAGAUGCUGGGCCGAUGGCCUUGGUGCUCGGCGCAAUGGCGCAAAAACAACAGCGGACGAAAUGGGACAUCCUGUGCUCCGGAAG .....((((((((((((((....((((.((((...)))).)))))))))...((.....((((....))))....)).)))))))))((.(((((....)))))..))...... (-39.24 = -39.04 + -0.20)



| Location | 10,480,078 – 10,480,194 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -44.78 |
| Consensus MFE | -43.82 |
| Energy contribution | -43.34 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10480078 116 - 22224390 UUCAGUUGAUGGCUCCUUGAGUGUAUGUAGCACUUCGGAGCUUCCGGAGCACAGGAUGUCCCAUUUCGUUCGCUGUUGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCC ....(((((((((..((((.((((.((((((.((((((.....))))))....(((((........)))))))))........((((....)))))).))))))))))))))))). ( -43.10) >DroSec_CAF1 10923 116 - 1 UUCAGUUGAUGGCUCCUUGAGUGUAUGUAGCACUUCGGAGCUUCCGGAGCACAGGAUGUCCCAUUUCGUCCACUGUUGUUCUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCC ....(((((((((..((((.((((.(((((((((((((.....))))))....(((((........)))))..))))).....((((....)))))).))))))))))))))))). ( -43.80) >DroSim_CAF1 11137 116 - 1 UUCAGUUGAUGGCUCCUUGAGUGUAUGUAGCACUUCGGAGCUUCCGGAGCACAGGAUGUCCCAUUUCGUCCACUGUUGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCC ....(((((((((..((((.((((.(((((((((((((.....))))))....(((((........)))))..))))).....((((....)))))).))))))))))))))))). ( -43.80) >DroEre_CAF1 12208 116 - 1 UUCAGCUGAUGGCUCCUUGAGUGUAUGUAGCACUUCGGAGUUUCCGGAGCACAGGAUGUCCCAUUUCGUCCGCUGUUGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCC ....(((((((((..((((.((((.((((((.((((((.....))))))....(((((........)))))))))........((((....)))))).))))))))))))))))). ( -48.50) >DroYak_CAF1 12756 116 - 1 UUCAGUUGAUGGCUCCUUGAGUGUAUGUAGCACUUCGGAGCUUCCGGAGCACAGGAUGUCCCAUUUCGUCCGCUGUUGUUUUUGCGCCAUAGCGUCGAGCACCAAGGCCAUCGGCC ....(((((((((..((((.((((...((((.((((((.....))))))....(((((........)))))))))......((((((....))).)))))))))))))))))))). ( -44.70) >consensus UUCAGUUGAUGGCUCCUUGAGUGUAUGUAGCACUUCGGAGCUUCCGGAGCACAGGAUGUCCCAUUUCGUCCGCUGUUGUUUUUGCGCCAUUGCGCCGAGCACCAAGGCCAUCGGCC ....(((((((((..((((.((((.(((((((((((((.....))))))....(((((........)))))..))))).....((((....)))))).))))))))))))))))). (-43.82 = -43.34 + -0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:52 2006