
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,477,791 – 10,478,149 |
| Length | 358 |
| Max. P | 0.991964 |

| Location | 10,477,791 – 10,477,909 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.793881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10477791 118 - 22224390 ACCCCAAAAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUACACC-CAAACCCCCCUGUUUAUACCUCAAACACCCCCGGGCUAUGCUAAUAACCAAUAUUUUAGA ..(((..(((((((.((((.......)))).)))))))....................-..........(((((.......))))).....))).....((((...........)))). ( -18.30) >DroSec_CAF1 8688 118 - 1 ACCCCAAAAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAACC-CCAGCCACCCUGUUUAUACCUCAAACACCCCCGGGCUAUGCUAAUAACCAAUAUUUUAGA .......(((((((....)....(((((((((......))))))..............-..((((....(((((.......)))))......))))..))).........))))))... ( -19.90) >DroSim_CAF1 8868 118 - 1 ACCCCAAAAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAACC-CCAGCCCCCCUGUUUAUACCUCAAACACCCCCGGGCUAUGCUAAUAACCAAUAUUUUAGA .......(((((((....)....(((((((((......))))))..............-..(((((...(((((.......))))).....)))))..))).........))))))... ( -22.30) >DroEre_CAF1 9950 118 - 1 ACCCCAAAAUGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAGCC-CAAACCCCCCUGUUUAUACCUCAAACACCCCCGGGCUAUGCUAAUAACCAAUAUUUUAGA .....(((((((.(....)....(((((((((......))))))..........((((-(.........(((((.......))))).....)))))..)))........)))))))... ( -21.74) >DroYak_CAF1 10517 119 - 1 ACCCCAAAAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUGUAAGCCCCAAACCCCCCUGUUUAUACCUCAAACACCCCCGGGCUAUGCUAAUAACCAAUAUUUUAGA .......(((((((.((((.......)))).))))))).............((((((((..........(((((.......))))).....))))).)))................... ( -21.16) >consensus ACCCCAAAAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAACC_CAAACCCCCCUGUUUAUACCUCAAACACCCCCGGGCUAUGCUAAUAACCAAUAUUUUAGA ..(((..(((((((.((((.......)))).)))))))............(((((((.............)))))))..............))).....((((...........)))). (-18.66 = -18.66 + 0.00)



| Location | 10,477,831 – 10,477,934 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10477831 103 + 22224390 GGUAUAAACAGGGGGGUUUG-GGUGUAUAAGGGAAGCUGACAAAGCAGCCAACGCUUUUUGUUGCCUACUUU-UUGGGGUCAGGGACUUGUUUUGUCAGCUUUCA--------------- .(((((..((((....))))-..)))))...(((((((((((((((((.(((((.....)))))(((((((.-...)))).)))...))))))))))))))))).--------------- ( -35.70) >DroSec_CAF1 8728 118 + 1 GGUAUAAACAGGGUGGCUGG-GGUUUAUAAGGGAAGCUGACAAAGCAGCCAACGCUUUUUGUUGCCUACUUU-UUGGGGUCAGGGACUUGUUUUGUCAGCUUUCAGUUGGGCGAGCGCAG ..(((((((.((....))..-.)))))))..(((((((((((((((((.(((((.....)))))(((((((.-...)))).)))...)))))))))))))))))......((....)).. ( -37.40) >DroSim_CAF1 8908 118 + 1 GGUAUAAACAGGGGGGCUGG-GGUUUAUAAGGGAAGCUGACAAAGCAGCCAACGCUUUUUGUUGCCUACUUU-UUGGGGUCAGGGACUUGUUUUGUCAGCUUUCAGUUGGGCGAGCGCAG .............(.(((..-.(((((....(((((((((((((((((.(((((.....)))))(((((((.-...)))).)))...)))))))))))))))))...))))).))).).. ( -37.70) >DroEre_CAF1 9990 114 + 1 GGUAUAAACAGGGGGGUUUG-GGCUUAUAAGGGAAGCUGACAAAGCAGCCAACGCUUUUUGUUGCCUACAUU-UUGGGGUCAGGGACUUGUUUUGUCAGCUUUCAGAGGGGCGG----CG .((.(((((......)))))-.((((.....(((((((((((((((((((.((.((...(((.....)))..-..)).))...)).).))))))))))))))))....)))).)----). ( -36.90) >DroYak_CAF1 10557 117 + 1 GGUAUAAACAGGGGGGUUUGGGGCUUACAAGGGAAGCUGACAAAGCAGCCAACGCUUUUUGUUGCCUACUUU-UUGGGGUCAGGGACUUGUUUUGUCAGCUUUCAGAGAGGCGAG--CAG .((.(((((......)))))..((((.....(((((((((((((((((.(((((.....)))))(((((((.-...)))).)))...)))))))))))))))))....))))..)--).. ( -37.40) >DroAna_CAF1 8414 97 + 1 GGGAGGAG-GGGGAGGUCUG----------GGGCAGCUGACAAAGCAGGCGACGCUUUUUGUUGCCUACUUUUUUGGGGUCAGGGACUUGUUUUGUCAGAGUAU------GCGG------ ....((..-(((.((((((.----------.(((..(..(.((((.((((((((.....)))))))).)))).)..).)))..)))))).)))..)).......------....------ ( -30.80) >consensus GGUAUAAACAGGGGGGUUUG_GGUUUAUAAGGGAAGCUGACAAAGCAGCCAACGCUUUUUGUUGCCUACUUU_UUGGGGUCAGGGACUUGUUUUGUCAGCUUUCAG__GGGCGA____AG ...............................(((((((((((((((((.(((((.....)))))(((((((.....)))).)))...)))))))))))))))))................ (-28.16 = -28.72 + 0.56)


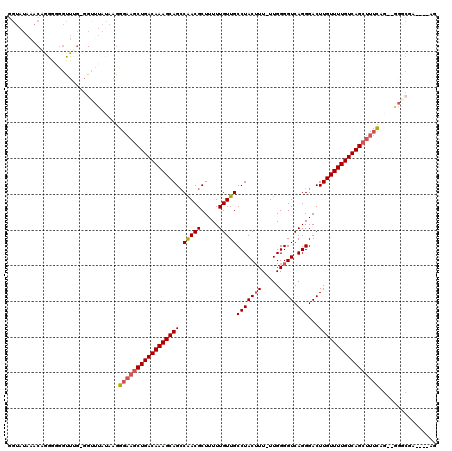
| Location | 10,477,831 – 10,477,934 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -21.85 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10477831 103 - 22224390 ---------------UGAAAGCUGACAAAACAAGUCCCUGACCCCAA-AAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUACACC-CAAACCCCCCUGUUUAUACC ---------------...((((((((.......(((...))).....-(((((((.((((.......)))).)))))))))))))))............-.................... ( -25.00) >DroSec_CAF1 8728 118 - 1 CUGCGCUCGCCCAACUGAAAGCUGACAAAACAAGUCCCUGACCCCAA-AAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAACC-CCAGCCACCCUGUUUAUACC ..((....))........((((((((.......(((...))).....-(((((((.((((.......)))).)))))))))))))))....(((((((.-...........))))))).. ( -27.60) >DroSim_CAF1 8908 118 - 1 CUGCGCUCGCCCAACUGAAAGCUGACAAAACAAGUCCCUGACCCCAA-AAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAACC-CCAGCCCCCCUGUUUAUACC ..((....))........((((((((.......(((...))).....-(((((((.((((.......)))).)))))))))))))))....(((((((.-...........))))))).. ( -27.60) >DroEre_CAF1 9990 114 - 1 CG----CCGCCCCUCUGAAAGCUGACAAAACAAGUCCCUGACCCCAA-AAUGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAGCC-CAAACCCCCCUGUUUAUACC ..----............(((((((((((....(((...))).....-...((((.((((.......)))).)))))))))))))))....(((((((.-...........))))))).. ( -26.40) >DroYak_CAF1 10557 117 - 1 CUG--CUCGCCUCUCUGAAAGCUGACAAAACAAGUCCCUGACCCCAA-AAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUGUAAGCCCCAAACCCCCCUGUUUAUACC ..(--((..(......(((.((((((.......(((...))).....-(((((((.((((.......)))).))))))))))))))))....)..)))...................... ( -28.00) >DroAna_CAF1 8414 97 - 1 ------CCGC------AUACUCUGACAAAACAAGUCCCUGACCCCAAAAAAGUAGGCAACAAAAAGCGUCGCCUGCUUUGUCAGCUGCCC----------CAGACCUCCCC-CUCCUCCC ------..((------(....(((((.......(((...)))......(((((((((.((.......)).)))))))))))))).)))..----------...........-........ ( -21.80) >consensus CU____UCGCCC__CUGAAAGCUGACAAAACAAGUCCCUGACCCCAA_AAAGUAGGCAACAAAAAGCGUUGGCUGCUUUGUCAGCUUCCCUUAUAAACC_CAAACCCCCCUGUUUAUACC ..................(((((((((((....(((...))).........((((.((((.......)))).)))))))))))))))................................. (-21.85 = -22.52 + 0.67)


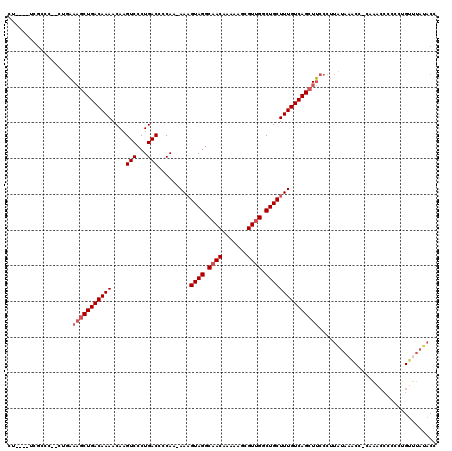
| Location | 10,477,934 – 10,478,040 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -26.24 |
| Energy contribution | -27.32 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10477934 106 - 22224390 GCGGAUAAAUAGAUGUAUAAGCGGGUUCUAUCUAUACAGAGUGCCCCUCCAUCAUACCACACUCGAUUCUGUGUGUGUCUGGCAAUUUGCUAGGCGCUCGCCCCUC---- ((((.......((((....((.((((....(((....)))..)))))).)))).....((((........))))(((((((((.....))))))))))))).....---- ( -31.60) >DroSec_CAF1 8846 108 - 1 GCGGAUAAAUAGAUGUAUAAGCGGGUUCUAUCUAUACAGAGUGCCCCUCCAUCAUACCUCACACGAUUCUGU--GUGUCUGGCAAUUUGCUAGGCGCUCGCCCCUCUGCU (((((......((((....((.((((....(((....)))..)))))).)))).................(.--(((((((((.....))))))))).).....))))). ( -31.60) >DroSim_CAF1 9026 108 - 1 GCGGAUAAAUAGAUGUAUAAGCGGGUUCUAUCUAUACAGAGUGCCCCUCCAUCAUACCCCACACGAUUCUGU--GUGUCUGGCAAUUUGCUAGGCGCUCGCCCCUCUGCU (((((......((((....((.((((....(((....)))..)))))).)))).................(.--(((((((((.....))))))))).).....))))). ( -31.60) >DroEre_CAF1 10104 106 - 1 GCGGAUAAAUAGAUGUAUAAGCGGGUUCUAUCUAUACAGAGUGCCCCUCCAUCAUACCCCACUCGAUUCUGU--GUGUCUGGCAAUUUGCUUGGCGCUCU--GCUCUGCU (((((....((((.((.(((((((((((((..((((((((((.......................)))))))--)))..))).))))))))))..)))))--).))))). ( -29.10) >DroYak_CAF1 10674 106 - 1 GCGGCUAAAUAGAUGUAUAAGCGGGUUCUAUCUAUACAAAGUGCCCCUCCAUCAUACCCCACUCGAUUCUGU--GUGUCUGGCAAAUCGCUAGGCGCUCU--GCUCCGCU ((((.......((((....((.((((.((..........)).)))))).)))).................(.--(((((((((.....))))))))).).--...)))). ( -29.30) >consensus GCGGAUAAAUAGAUGUAUAAGCGGGUUCUAUCUAUACAGAGUGCCCCUCCAUCAUACCCCACUCGAUUCUGU__GUGUCUGGCAAUUUGCUAGGCGCUCGCCCCUCUGCU (((((......((((....((.((((....(((....)))..)))))).)))).....................(((((((((.....))))))))).......))))). (-26.24 = -27.32 + 1.08)

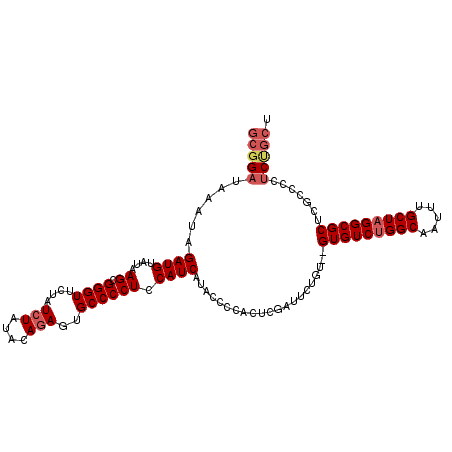
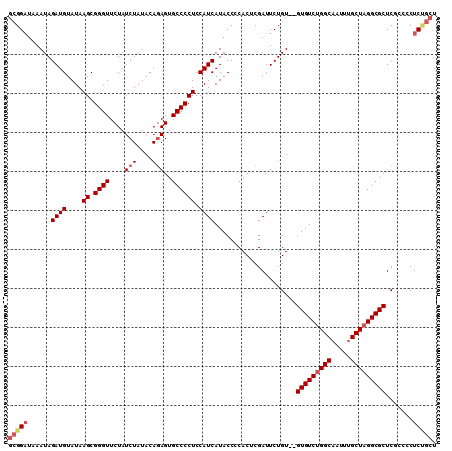
| Location | 10,478,040 – 10,478,149 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.71 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10478040 109 + 22224390 UACCCACAAGUUUUAUUCAGCCCCCUGCUGCCGCCGCUGCAUCUUUUGUAUUGUGAAUCAUAGC-CAAAUGCAUUCGACGCCAUGCCAUCCCAUGGCCAAUGGGGCUGAC ................((((((((.(((.((....)).)))........((((((((((((...-...))).)))))..((((((......)))))))))))))))))). ( -32.70) >DroSec_CAF1 8954 109 + 1 UACCCACAAGUUUUAUUCAGCC-CCUGCUGCCGCCGCUGCAUCUUUUGUAUUGUGAAUCGUAGCCCAAAUGCAUUCGACGCCAUGCCAUCCCAUGGCCAAUGGGGCUGAC ................((((((-((.(((((...(((((((.....))))..)))....)))))...............((((((......))))))....)))))))). ( -33.90) >DroSim_CAF1 9134 109 + 1 UACCCACAAGUUUUAUUCAGCCCCCUGCUGCCGCCGCUGCAUCUUUUGUAUUGUGAAUCAUAGC-CAAAUGCAUUCGACGCCAUGCCAUCCCAUGGCCAAUGAGGCUGAC ................((((((...(((.((....)).))).......(((((((((((((...-...))).)))))..((((((......))))))))))).)))))). ( -27.40) >DroEre_CAF1 10210 109 + 1 UACCCACAAGUUUUACUUAACCCCCUGCUGCCGCCGCUGCAUCUUUUGUAUUGUGAAUCAUAGC-CAAAUGCAUUCGACGCCAUGCCAUCCCAUGGCCAAUGGGGCUGAC ................(((.((((.(((.((....)).)))........((((((((((((...-...))).)))))..((((((......)))))))))))))).))). ( -23.80) >DroYak_CAF1 10780 109 + 1 UACCCACAAGUUUUAUUUAAACCCCUGCUGCCGCCGCUGCAUCUUUUGUAUUGUGAAUCAUAGC-CAAAUGCAUUCGACGCCAUGCCAUCCCAUGGCCAAUGGGGCUGAC .....................(((((((.((....)).)))........((((((((((((...-...))).)))))..((((((......))))))))))))))..... ( -23.80) >consensus UACCCACAAGUUUUAUUCAGCCCCCUGCUGCCGCCGCUGCAUCUUUUGUAUUGUGAAUCAUAGC_CAAAUGCAUUCGACGCCAUGCCAUCCCAUGGCCAAUGGGGCUGAC ................((((((((.(((.((....)).)))........((((((((((((.......))).)))))..((((((......)))))))))))))))))). (-25.12 = -25.72 + 0.60)

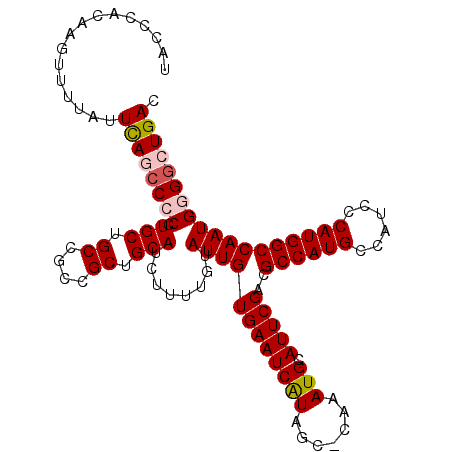
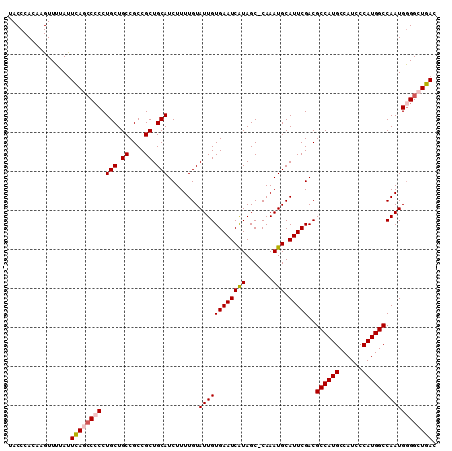
| Location | 10,478,040 – 10,478,149 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.71 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -34.12 |
| Energy contribution | -34.76 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10478040 109 - 22224390 GUCAGCCCCAUUGGCCAUGGGAUGGCAUGGCGUCGAAUGCAUUUG-GCUAUGAUUCACAAUACAAAAGAUGCAGCGGCGGCAGCAGGGGGCUGAAUAAAACUUGUGGGUA .((((((((.(((((((((......))))))((((..(((((((.-(.(((........))))...)))))))....))))..)))))))))))................ ( -40.90) >DroSec_CAF1 8954 109 - 1 GUCAGCCCCAUUGGCCAUGGGAUGGCAUGGCGUCGAAUGCAUUUGGGCUACGAUUCACAAUACAAAAGAUGCAGCGGCGGCAGCAGG-GGCUGAAUAAAACUUGUGGGUA .((((((((....((((((......))))))((((..(((((((.(................)...)))))))....))))....))-))))))................ ( -39.09) >DroSim_CAF1 9134 109 - 1 GUCAGCCUCAUUGGCCAUGGGAUGGCAUGGCGUCGAAUGCAUUUG-GCUAUGAUUCACAAUACAAAAGAUGCAGCGGCGGCAGCAGGGGGCUGAAUAAAACUUGUGGGUA .((((((((.(((((((((......))))))((((..(((((((.-(.(((........))))...)))))))....))))..)))))))))))................ ( -37.90) >DroEre_CAF1 10210 109 - 1 GUCAGCCCCAUUGGCCAUGGGAUGGCAUGGCGUCGAAUGCAUUUG-GCUAUGAUUCACAAUACAAAAGAUGCAGCGGCGGCAGCAGGGGGUUAAGUAAAACUUGUGGGUA ...((((((.(((((((((......))))))((((..(((((((.-(.(((........))))...)))))))....))))..))))))))).................. ( -34.00) >DroYak_CAF1 10780 109 - 1 GUCAGCCCCAUUGGCCAUGGGAUGGCAUGGCGUCGAAUGCAUUUG-GCUAUGAUUCACAAUACAAAAGAUGCAGCGGCGGCAGCAGGGGUUUAAAUAAAACUUGUGGGUA ((((..(((((.....))))).)))).((.(((((..(((((((.-(.(((........))))...))))))).))))).)).((.((((((.....)))))).)).... ( -33.30) >consensus GUCAGCCCCAUUGGCCAUGGGAUGGCAUGGCGUCGAAUGCAUUUG_GCUAUGAUUCACAAUACAAAAGAUGCAGCGGCGGCAGCAGGGGGCUGAAUAAAACUUGUGGGUA .((((((((.(((((((((......))))))((((..(((((((....(((........)))....)))))))....))))..)))))))))))................ (-34.12 = -34.76 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:49 2006