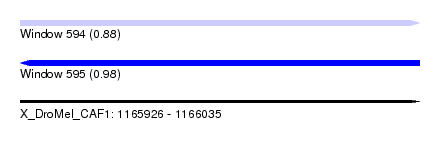
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,165,926 – 1,166,035 |
| Length | 109 |
| Max. P | 0.984522 |

| Location | 1,165,926 – 1,166,035 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.16 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
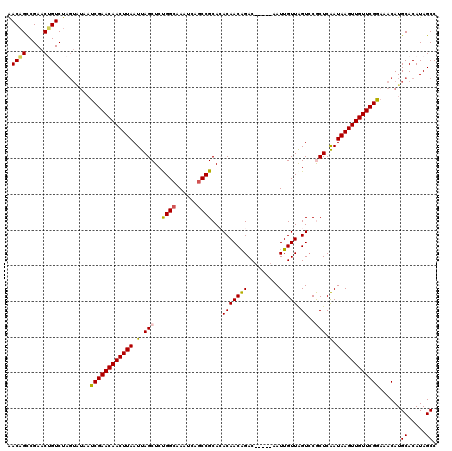
>X_DroMel_CAF1 1165926 109 + 22224390 GACAGCCGAACUGUCUA------UCGAACAACUUAAUUAGCUCUGGCAAAUCAGCCGCACACAACAGAC-----AAUUGUUAGUCCGCUCGAUAAGUUGUUCGGCAACAUGCACAUAGCC (((((.....)))))..------.(((((((((((.(.(((...(((......)))..........(((-----........))).))).).)))))))))))((.....))........ ( -30.60) >DroSec_CAF1 1995 115 + 1 AACAGUCGAACUGUCUAGUGUAAUCGAACAACUUAAUUAGCUCCGGAAAAUCAGCCGCACACAACAGAC-----AAUUGUUAGUCCGCUCAAUAAGUUGUUCGGAAACAUGCACAUAGCC .((((.....))))((((((((...((((((((((.(.(((..(((........))).........(((-----........))).))).).))))))))))(....).))))).))).. ( -30.00) >DroSim_CAF1 2598 115 + 1 AACAGUCGAACUGUCUAGUGUAAUCGAACAACUUAAUUAGCUCCGGCAAAUCAGCCGCACACAACAGAC-----AAUUGUUAGUCCGCUCAAUAAGUUGUUCGGAAACAUGCACAUAGCC .((((.....))))((((((((...((((((((((.(.(((..((((......)))).........(((-----........))).))).).))))))))))(....).))))).))).. ( -33.80) >DroEre_CAF1 1041 120 + 1 AACUGCCGAACAGUCUAGUAUAAUCGAACAACUUAAUUAGCUCUGGCAAAUCAGCCGCAAACAACGGACAAUACAAUUGUUAGUCCACUCAAUAAGUUGUUCGGAAACAUGCACAUAGCC .((((.....))))...((((....((((((((((....((...(((......))))).......((((((((....)))).))))......))))))))))(....)))))........ ( -30.00) >DroYak_CAF1 3238 115 + 1 AACAGCCGAACAGUCUAGUAUGAUCGAACAACUUAAUUAGCUCUGGCAAAUGAGCCGCAAACAACGGAC-----AAUUGUUGGUGCACUCGAUAAGUUGUUCGAAAACAUGCACAUAGCC .................(((((.((((((((((((....((.(..((((.((..(((.......))).)-----).))))..).))......))))))))))))...)))))........ ( -33.20) >consensus AACAGCCGAACUGUCUAGUAUAAUCGAACAACUUAAUUAGCUCUGGCAAAUCAGCCGCACACAACAGAC_____AAUUGUUAGUCCGCUCAAUAAGUUGUUCGGAAACAUGCACAUAGCC .((((.....)))).........((((((((((((.(.(((..((((......))))...(((((((.........))))).))..))).).))))))))))))......((.....)). (-21.48 = -21.16 + -0.32)

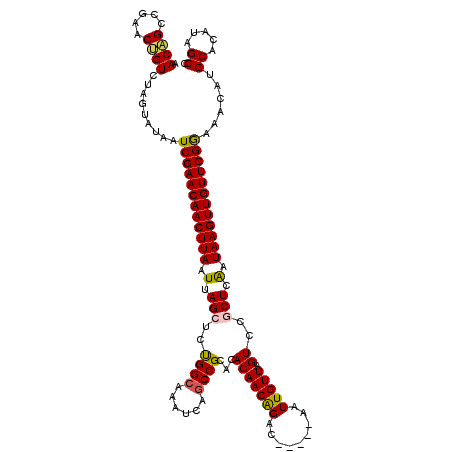
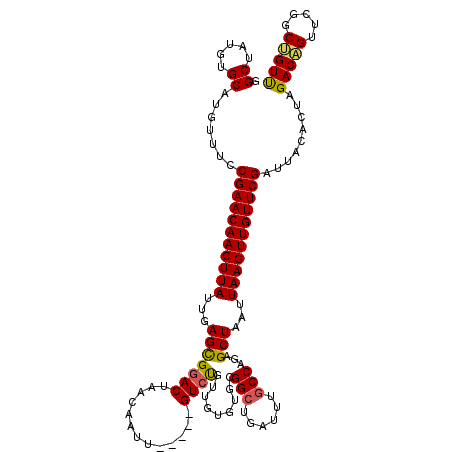
| Location | 1,165,926 – 1,166,035 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -29.84 |
| Energy contribution | -29.56 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1165926 109 - 22224390 GGCUAUGUGCAUGUUGCCGAACAACUUAUCGAGCGGACUAACAAUU-----GUCUGUUGUGUGCGGCUGAUUUGCCAGAGCUAAUUAAGUUGUUCGA------UAGACAGUUCGGCUGUC (((.((......)).)))((((((((((...((((.(((((((...-----...))))).)).)(((......)))...)))...))))))))))..------..(((((.....))))) ( -37.90) >DroSec_CAF1 1995 115 - 1 GGCUAUGUGCAUGUUUCCGAACAACUUAUUGAGCGGACUAACAAUU-----GUCUGUUGUGUGCGGCUGAUUUUCCGGAGCUAAUUAAGUUGUUCGAUUACACUAGACAGUUCGACUGUU .((.....)).(((...(((((((((((...(((((((........-----)))))))....((..(((......))).))....)))))))))))...)))...(((((.....))))) ( -33.00) >DroSim_CAF1 2598 115 - 1 GGCUAUGUGCAUGUUUCCGAACAACUUAUUGAGCGGACUAACAAUU-----GUCUGUUGUGUGCGGCUGAUUUGCCGGAGCUAAUUAAGUUGUUCGAUUACACUAGACAGUUCGACUGUU .((.....)).(((...(((((((((((...(((((((........-----))))........((((......))))..)))...)))))))))))...)))...(((((.....))))) ( -37.30) >DroEre_CAF1 1041 120 - 1 GGCUAUGUGCAUGUUUCCGAACAACUUAUUGAGUGGACUAACAAUUGUAUUGUCCGUUGUUUGCGGCUGAUUUGCCAGAGCUAAUUAAGUUGUUCGAUUAUACUAGACUGUUCGGCAGUU .((.....)).......(((((((((((...(((((((..((....))...)))).........(((......)))...)))...))))))))))).........(((((.....))))) ( -34.30) >DroYak_CAF1 3238 115 - 1 GGCUAUGUGCAUGUUUUCGAACAACUUAUCGAGUGCACCAACAAUU-----GUCCGUUGUUUGCGGCUCAUUUGCCAGAGCUAAUUAAGUUGUUCGAUCAUACUAGACUGUUCGGCUGUU ((((....(((.((((((((((((((((...((((((..((((((.-----....)))))))))(((......)))...)))...)))))))))))).......)))))))..))))... ( -34.91) >consensus GGCUAUGUGCAUGUUUCCGAACAACUUAUUGAGCGGACUAACAAUU_____GUCUGUUGUGUGCGGCUGAUUUGCCAGAGCUAAUUAAGUUGUUCGAUUACACUAGACAGUUCGGCUGUU .((.....)).......(((((((((((...(((((((.............)))).........(((......)))...)))...))))))))))).........(((((.....))))) (-29.84 = -29.56 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:23 2006