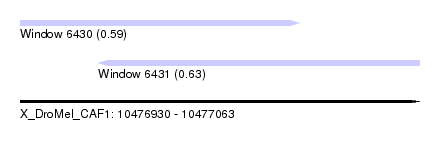
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,476,930 – 10,477,063 |
| Length | 133 |
| Max. P | 0.634027 |

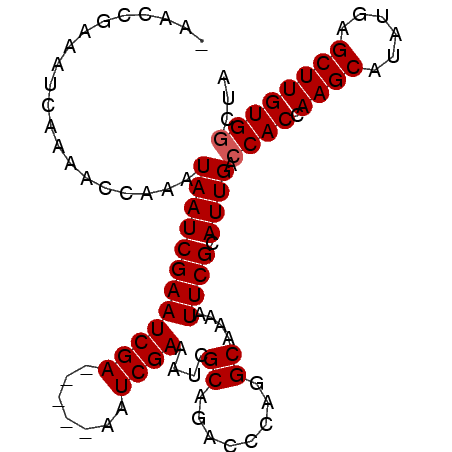
| Location | 10,476,930 – 10,477,023 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -17.07 |
| Consensus MFE | -14.85 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10476930 93 + 22224390 -AACCGAAAUCAAAACCAAAUAAUCGAAUCGA-----AAUCGAAAUCGCAGACCCAGGCAAAAUUCGCAUUGACCACCAAGCAUAUGAGCUUGUGGCUA -...................(((((((((((.-----...)))....((........))....)))).)))).((((.((((......))))))))... ( -16.10) >DroSec_CAF1 7788 93 + 1 -AACCGAAAUCAAAACCAAAUAAUCGAAUCGA-----AAUCGAAAUCGCAGACCCAUGCAAAAUUCGCAUUGACCACCAAGCAUAUGAGCUUGUGGCUA -...................(((((((((((.-----...)))....(((......)))....)))).)))).((((.((((......))))))))... ( -18.10) >DroSim_CAF1 7974 93 + 1 -AACCGAAAUCAAAACCAAAUAAUCGAAUCGA-----AAUCGAAAUCGCAGACCCAGGCAAAAUUCGCAUUGAGCACCAAGCAUAUGAGCUUGUGGCUA -...................(((((((((((.-----...)))....((........))....)))).))))(((..(((((......)))))..))). ( -17.30) >DroEre_CAF1 9026 99 + 1 AAACCAAAAUCAAAACCAAAUAAUCGAAUCGAAUCGAAAUCGAAAUCGCAGACCCAGGCAAAAUUCGCAUUGACCACCAAGCAUAUGAGCUUGUGGCUA .......................((((..(((((((....)))....((........))....))))..))))((((.((((......))))))))... ( -16.80) >consensus _AACCGAAAUCAAAACCAAAUAAUCGAAUCGA_____AAUCGAAAUCGCAGACCCAGGCAAAAUUCGCAUUGACCACCAAGCAUAUGAGCUUGUGGCUA ....................((((((((((((.......))))....((........))....)))).)))).((((.((((......))))))))... (-14.85 = -15.10 + 0.25)


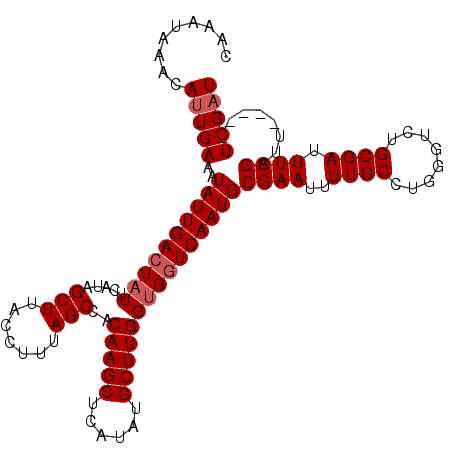
| Location | 10,476,956 – 10,477,063 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.53 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10476956 107 - 22224390 CAAAUAAACAUUGAAAUAUUGACUAUUCAUAGCUUACCUUUAGCCACAAGCUCAUAUGCUUGGUGGUCAAUGCGAAUUUUGCCUGGGUCUGCGAUUUCGAUU-----UCGAU .........(((((((((((((((((.....(((.......)))..(((((......)))))))))))))).((((..((((........)))).)))))))-----))))) ( -27.80) >DroSec_CAF1 7814 107 - 1 CAAAUAAACAUUGAAAUAUUGACUAUUCAUAGCUUACCUUUAGCCACAAGCUCAUAUGCUUGGUGGUCAAUGCGAAUUUUGCAUGGGUCUGCGAUUUCGAUU-----UCGAU .........(((((((((((((((((.....(((.......)))..(((((......)))))))))))))).((((..(((((......))))).)))))))-----))))) ( -29.60) >DroSim_CAF1 8000 107 - 1 CAAAUAAACAUUGAAAUAUUGACUAUUCAUAGCUUACCUUUAGCCACAAGCUCAUAUGCUUGGUGCUCAAUGCGAAUUUUGCCUGGGUCUGCGAUUUCGAUU-----UCGAU .........((((((((..(((....)))..((..((((...((..(((((......))))).(((.....)))......))..))))..)).)))))))).-----..... ( -22.30) >DroEre_CAF1 9053 112 - 1 CAAAUAAACACUGAAAUAUUGACUUUUCAUAGCUUACCUUUAGCCACAAGCUCAUAUGCUUGGUGGUCAAUGCGAAUUUUGCCUGGGUCUGCGAUUUCGAUUUCGAUUCGAU ...........(((((........)))))..((..((((...(((((((((......)))).)))))....((((...))))..))))..))((..(((....))).))... ( -24.20) >consensus CAAAUAAACAUUGAAAUAUUGACUAUUCAUAGCUUACCUUUAGCCACAAGCUCAUAUGCUUGGUGGUCAAUGCGAAUUUUGCCUGGGUCUGCGAUUUCGAUU_____UCGAU .........(((((..((((((((((.....(((.......)))..(((((......)))))))))))))))((((..((((........)))).))))........))))) (-22.77 = -23.53 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:43 2006