
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,474,343 – 10,474,475 |
| Length | 132 |
| Max. P | 0.908787 |

| Location | 10,474,343 – 10,474,439 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10474343 96 + 22224390 CUGAAU-CAAUGAUAUUAAGUAAAGCAGAUGUCACGGAUGUAGUGCA-GUUAAGGGUUAACCUUAACAU-----------CAUCCCGCUGUUGCGUUAACAGCGCGUAA (((.((-(..(((((((..........)))))))..))).))).((.-(((((((.....)))))))..-----------......(((((((...))))))))).... ( -27.40) >DroSec_CAF1 5134 95 + 1 CUGAAUCCAGAGAUC-UGAGUGUAGCAGAUGUCACGGAUGUAGUGCA-GUUAAGGGUUAACCUUAACAU-----------CAUCCCGCUGUUGUGUUAACAGCGC-UAA (((.((((.((.(((-((.......))))).))..)))).))).((.-(((((((.....)))))))..-----------......(((((((...)))))))))-... ( -31.40) >DroSim_CAF1 5325 94 + 1 CUGAAU-UAGAGAUA-UGAGUGUAGCAGAUGUCACGGAUGCAGUGCA-GUUAAGGGUUAACCUUAACAU-----------CAUCCCGCUGUUGUGUUAACAGCGC-UAA ......-........-......((((.((((.(((.......)))..-(((((((.....)))))))..-----------))))..(((((((...)))))))))-)). ( -25.40) >DroEre_CAF1 6329 108 + 1 CUAAAU-CAAAACUAUUAAAUAUUGCAGAUGUCCCGGAUGAAGUGCAUGUUAAGGGUUAACCUUAACAUAGCCGCUGUUUAAUCCCGCUGUUGCGUUAACAGCACGUAA ......-...............((((.........((((..((((((((((((((.....))))))))).).)))).....)))).(((((((...)))))))..)))) ( -28.70) >DroYak_CAF1 6847 107 + 1 CUGAAU-CAGUGAUAUUAAAUAUUGCAGAUGUCACAGAUAAAGUGCA-GUUAAGGGUUAACCUUAACAUAGCCGCUGGUUCAUCCUGCUGUUGCGUUAACAGCGCGUAA .(((((-(((((...........((((..((((...))))...))))-(((((((.....))))))).....))))))))))....(((((((...)))))))...... ( -33.50) >consensus CUGAAU_CAGAGAUAUUAAGUAUAGCAGAUGUCACGGAUGAAGUGCA_GUUAAGGGUUAACCUUAACAU___________CAUCCCGCUGUUGCGUUAACAGCGCGUAA ........................((.((((.(((.......)))...(((((((.....))))))).............))))..(((((((...))))))))).... (-18.88 = -19.48 + 0.60)

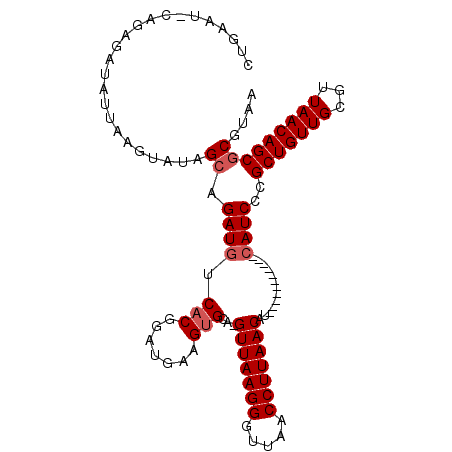

| Location | 10,474,343 – 10,474,439 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10474343 96 - 22224390 UUACGCGCUGUUAACGCAACAGCGGGAUG-----------AUGUUAAGGUUAACCCUUAAC-UGCACUACAUCCGUGACAUCUGCUUUACUUAAUAUCAUUG-AUUCAG .....(((((((.....))))))).((((-----------((((((((((((.....))))-(((((.......))).)).........)))))))))))).-...... ( -24.70) >DroSec_CAF1 5134 95 - 1 UUA-GCGCUGUUAACACAACAGCGGGAUG-----------AUGUUAAGGUUAACCCUUAAC-UGCACUACAUCCGUGACAUCUGCUACACUCA-GAUCUCUGGAUUCAG .((-((((((((.....)))))).(((((-----------..(((((((.....)))))))-..(((.......))).)))))))))...(((-(....))))...... ( -27.80) >DroSim_CAF1 5325 94 - 1 UUA-GCGCUGUUAACACAACAGCGGGAUG-----------AUGUUAAGGUUAACCCUUAAC-UGCACUGCAUCCGUGACAUCUGCUACACUCA-UAUCUCUA-AUUCAG .((-((((((((.....)))))).(((((-----------..(((((((.....)))))))-..(((.......))).)))))))))......-........-...... ( -25.20) >DroEre_CAF1 6329 108 - 1 UUACGUGCUGUUAACGCAACAGCGGGAUUAAACAGCGGCUAUGUUAAGGUUAACCCUUAACAUGCACUUCAUCCGGGACAUCUGCAAUAUUUAAUAGUUUUG-AUUUAG .....(((((((..(((....)))......)))))))((.(((((((((.....)))))))))))........((((((.................))))))-...... ( -24.83) >DroYak_CAF1 6847 107 - 1 UUACGCGCUGUUAACGCAACAGCAGGAUGAACCAGCGGCUAUGUUAAGGUUAACCCUUAAC-UGCACUUUAUCUGUGACAUCUGCAAUAUUUAAUAUCACUG-AUUCAG ...(((((((((.....)))))).((.....)).)))((...(((((((.....)))))))-.)).((..(((.((((..................)))).)-))..)) ( -26.37) >consensus UUACGCGCUGUUAACGCAACAGCGGGAUG___________AUGUUAAGGUUAACCCUUAAC_UGCACUACAUCCGUGACAUCUGCUAUACUUAAUAUCUCUG_AUUCAG ....((((((((.....)))))).(((((.............(((((((.....)))))))...(((.......))).)))))))........................ (-19.32 = -19.56 + 0.24)



| Location | 10,474,379 – 10,474,475 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10474379 96 + 22224390 AUGUAGUGCA-GUUAAGGGUUAACCUUAACAU-----------CAUCCCGCUGUUGCGUUAACAGCGCGUAACGGGACUGCCGCUCUCUUUGUCCAG-CCCCCCUC--CGA ..((..(((.-(((((((.....)))))))..-----------......(((((((...))))))))))..))(((.(((..((.......)).)))-))).....--... ( -28.50) >DroSec_CAF1 5170 97 + 1 AUGUAGUGCA-GUUAAGGGUUAACCUUAACAU-----------CAUCCCGCUGUUGUGUUAACAGCGC-UAACGGGACUGCCGCUCUCUUUGUCAAU-CCCCCCUCACCAA ..((...((.-(((((((.....)))))))..-----------......(((((((...)))))))))-..))((((.((.((.......)).)).)-))).......... ( -25.10) >DroSim_CAF1 5360 97 + 1 AUGCAGUGCA-GUUAAGGGUUAACCUUAACAU-----------CAUCCCGCUGUUGUGUUAACAGCGC-UAACGGGACUGCCGCUCUCUUUGUCAAU-CCCCCCUCACCGA ..(((((...-(((((((.....)))))))..-----------...((((.((((((....)))))).-...)))))))))................-............. ( -26.80) >DroEre_CAF1 6365 110 + 1 AUGAAGUGCAUGUUAAGGGUUAACCUUAACAUAGCCGCUGUUUAAUCCCGCUGUUGCGUUAACAGCACGUAACGGGACUGCCGCGCUCUCUU-UUGUGUCCCCCUCUCUCU .....(.(((((((((((.....))))))))).)))((((((......(((....)))..)))))).......(((((..............-....)))))......... ( -31.57) >DroYak_CAF1 6883 110 + 1 AUAAAGUGCA-GUUAAGGGUUAACCUUAACAUAGCCGCUGGUUCAUCCUGCUGUUGCGUUAACAGCGCGUAACGGGACUGCCGCUCUCUUUGCCCGUCUUCCCCUCUCUCA .(((((.((.-(((((((.....))))))).........(((...(((((....(((((.....)))))...)))))..)))))...)))))................... ( -29.80) >consensus AUGAAGUGCA_GUUAAGGGUUAACCUUAACAU___________CAUCCCGCUGUUGCGUUAACAGCGCGUAACGGGACUGCCGCUCUCUUUGUCAAU_CCCCCCUCACCCA ....((.((..(((((((.....)))))))...............(((((....(((((.....)))))...))))).....)).))........................ (-22.54 = -22.82 + 0.28)

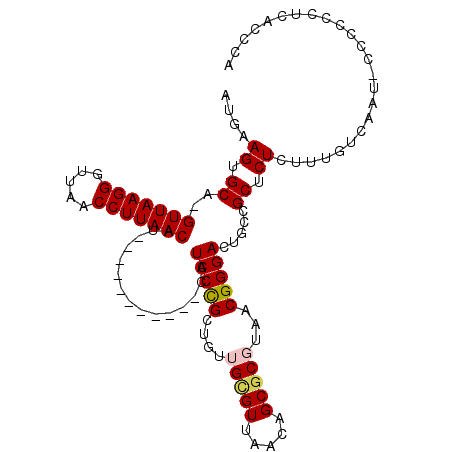

| Location | 10,474,379 – 10,474,475 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Mean single sequence MFE | -33.09 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10474379 96 - 22224390 UCG--GAGGGGGG-CUGGACAAAGAGAGCGGCAGUCCCGUUACGCGCUGUUAACGCAACAGCGGGAUG-----------AUGUUAAGGUUAACCCUUAAC-UGCACUACAU ...--....((((-(((..............)))))))(((((.(((((((.....))))))).).))-----------))(((((((.....)))))))-.......... ( -30.44) >DroSec_CAF1 5170 97 - 1 UUGGUGAGGGGGG-AUUGACAAAGAGAGCGGCAGUCCCGUUA-GCGCUGUUAACACAACAGCGGGAUG-----------AUGUUAAGGUUAACCCUUAAC-UGCACUACAU .(((((((.((((-.(((((......((((...((((((...-.)((((((.....))))))))))).-----------.))))...)))))))))...)-).)))))... ( -32.90) >DroSim_CAF1 5360 97 - 1 UCGGUGAGGGGGG-AUUGACAAAGAGAGCGGCAGUCCCGUUA-GCGCUGUUAACACAACAGCGGGAUG-----------AUGUUAAGGUUAACCCUUAAC-UGCACUGCAU .(((((((.((((-.(((((......((((...((((((...-.)((((((.....))))))))))).-----------.))))...)))))))))...)-).)))))... ( -33.60) >DroEre_CAF1 6365 110 - 1 AGAGAGAGGGGGACACAA-AAGAGAGCGCGGCAGUCCCGUUACGUGCUGUUAACGCAACAGCGGGAUUAAACAGCGGCUAUGUUAAGGUUAACCCUUAACAUGCACUUCAU .....((((.........-.......(((...(((((((((...(((.......)))..))))))))).....)))((.(((((((((.....))))))))))).)))).. ( -36.20) >DroYak_CAF1 6883 110 - 1 UGAGAGAGGGGAAGACGGGCAAAGAGAGCGGCAGUCCCGUUACGCGCUGUUAACGCAACAGCAGGAUGAACCAGCGGCUAUGUUAAGGUUAACCCUUAAC-UGCACUUUAU ....((((((((...((..(.....)..))....))))....(((((((((.....)))))).((.....)).)))((...(((((((.....)))))))-.)).)))).. ( -32.30) >consensus UCGGAGAGGGGGG_AUGGACAAAGAGAGCGGCAGUCCCGUUACGCGCUGUUAACGCAACAGCGGGAUG___________AUGUUAAGGUUAACCCUUAAC_UGCACUACAU ..............................(((((((((.....)((((((.....)))))))))))..............(((((((.....))))))).)))....... (-21.66 = -21.70 + 0.04)

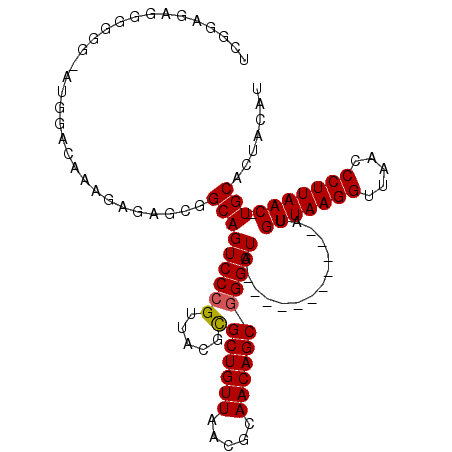

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:39 2006