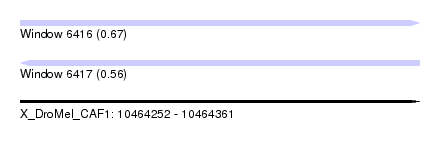
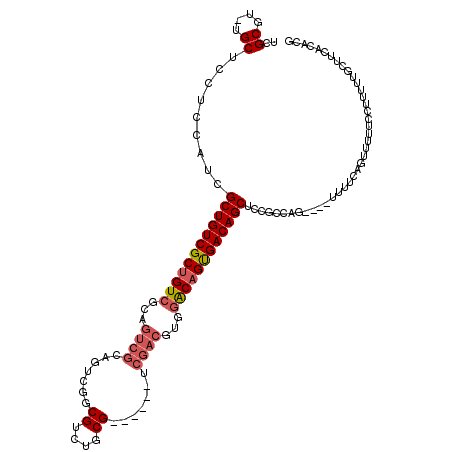
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,464,252 – 10,464,361 |
| Length | 109 |
| Max. P | 0.669212 |

| Location | 10,464,252 – 10,464,361 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -26.26 |
| Energy contribution | -28.14 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10464252 109 + 22224390 CGUGUGAAGCAAAAAGGAAAAACUGAAAA----CUGACAGAGCUGUCACUGCCAACGUCGA------CGCAGACGCCGACUGCGACUGCGACAGCGACAGCGAUGGAGGCGCU-ACGCGA (((((..(((...................----.(((((....)))))..(((..((((..------(((((.(((.....))).)))))...((....))))))..))))))-))))). ( -38.20) >DroSec_CAF1 8558 109 + 1 CGUGUGAAGCAAAAAGGAAAAACUGAAAA----CUGGCGGAGCUGUCACUGUCCACGUCGA------CGCAGACGCCGACUGCGACUGCGACAGCGACAGCGAUGGAGGAGCA-ACGCGA (((((...((....((......)).....----((..((..((((((.(((((...(....------)((((.(((.....))).))))))))).))))))..))..)).)).-))))). ( -38.20) >DroSim_CAF1 8529 103 + 1 CGUGUGAAGCAAAAAGGAAAAACUGAAAA----CUGGCGGAGCUGUCACUGUCCACCUCGA------CGCCGACGCCGACU------GCGACAGCGACAGCGAUGGAGGAGCA-ACGCGA (((((...((....((......)).....----((..((..((((((.(((((((..(((.------((....)).))).)------).))))).))))))..))..)).)).-))))). ( -33.50) >DroYak_CAF1 8768 120 + 1 CGUUUGGGCCAAAAAGGAAAAACUGAAUAUCUGCUGACGGAGCUGUCGCUGUACACGUCGACGUAGACGCAGACGCCGACGCCGACGCCGACUGCGACAGCGACGGAGGGGCAAACGCGA (((.(..(((....((......)).........((..((..((((((((.((...(((((.(((....((....))..))).)))))...)).))))))))..))..)))))..).))). ( -44.80) >consensus CGUGUGAAGCAAAAAGGAAAAACUGAAAA____CUGACGGAGCUGUCACUGUCCACGUCGA______CGCAGACGCCGACUGCGACUGCGACAGCGACAGCGAUGGAGGAGCA_ACGCGA (((((...((....((......)).........((..((..((((((.(((((...((((........((....))......))))...))))).))))))..))..)).))..))))). (-26.26 = -28.14 + 1.88)

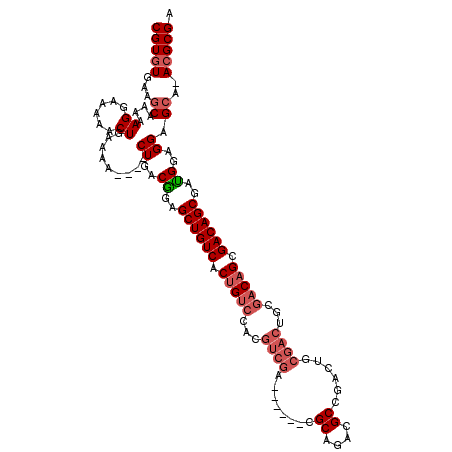

| Location | 10,464,252 – 10,464,361 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -23.92 |
| Energy contribution | -25.04 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10464252 109 - 22224390 UCGCGU-AGCGCCUCCAUCGCUGUCGCUGUCGCAGUCGCAGUCGGCGUCUGCG------UCGACGUUGGCAGUGACAGCUCUGUCAG----UUUUCAGUUUUUCCUUUUUGCUUCACACG ..(.(.-(((.........((((((((((((((....)).(((((((....))------)))))...)))))))))))).(((....----....)))............))).).)... ( -40.80) >DroSec_CAF1 8558 109 - 1 UCGCGU-UGCUCCUCCAUCGCUGUCGCUGUCGCAGUCGCAGUCGGCGUCUGCG------UCGACGUGGACAGUGACAGCUCCGCCAG----UUUUCAGUUUUUCCUUUUUGCUUCACACG ..(((.-((......))..((((((((((((((....)).(((((((....))------)))))...))))))))))))..)))...----............................. ( -37.60) >DroSim_CAF1 8529 103 - 1 UCGCGU-UGCUCCUCCAUCGCUGUCGCUGUCGC------AGUCGGCGUCGGCG------UCGAGGUGGACAGUGACAGCUCCGCCAG----UUUUCAGUUUUUCCUUUUUGCUUCACACG ..(((.-((......))..((((((((((((((------..((((((....))------)))).)).))))))))))))..)))...----............................. ( -33.60) >DroYak_CAF1 8768 120 - 1 UCGCGUUUGCCCCUCCGUCGCUGUCGCAGUCGGCGUCGGCGUCGGCGUCUGCGUCUACGUCGACGUGUACAGCGACAGCUCCGUCAGCAGAUAUUCAGUUUUUCCUUUUUGGCCCAAACG ....((((((.....((..((((((((.((..((((((((((.((((....)))).))))))))))..)).))))))))..))...)))))).....((((..((.....))...)))). ( -49.90) >consensus UCGCGU_UGCUCCUCCAUCGCUGUCGCUGUCGCAGUCGCAGUCGGCGUCUGCG______UCGACGUGGACAGUGACAGCUCCGCCAG____UUUUCAGUUUUUCCUUUUUGCUUCACACG ..((....)).........((((((((((((...((((.......((....)).......))))...))))))))))))......................................... (-23.92 = -25.04 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:31 2006