
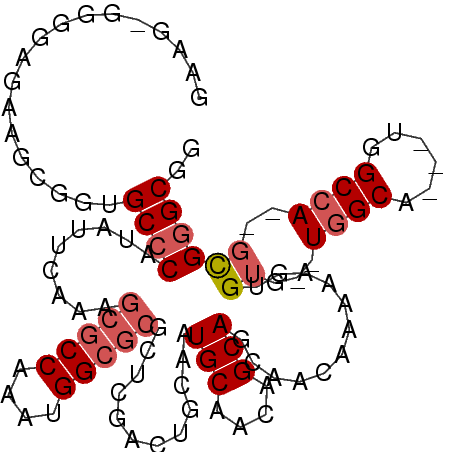
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,460,190 – 10,460,281 |
| Length | 91 |
| Max. P | 0.937665 |

| Location | 10,460,190 – 10,460,281 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -19.89 |
| Energy contribution | -21.20 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10460190 91 + 22224390 GAAGCGGGGAAUGGCGGUGCCCAUAUUCAAAGCGCCAAAUGG------------CAAUGCAACAGCAGCAACAAAAAGUGUG--UGGCCCGCUGGCCA---UGGGCGA ...((..((...(((((.(((((((((....(((((....))------------)..(((....))))).......))))))--.))))))))..)).---...)).. ( -31.00) >DroSec_CAF1 4660 101 + 1 GAAGAGGGGAGAAGCGGUGCCCAUAUUCAAAGCGCCAAAUGGCGCGCUCGACUGCAAUGCAACAGCAGCAACAAAAAGUGUG--UGGCA---UGGCCA--GCGGGCGG (((...(((..........)))...)))...(((((....)))))(((((.((((.((((.(((...((........)).))--).)))---).).))--)))))).. ( -35.40) >DroSim_CAF1 4428 101 + 1 NNNNNNNNNNNNNNNNNNGCCCAUAUUCAAAGCGCCAAAUGGCGCGCUCGACUGCAAUGCAACAGCAGCAACAAAAAGUGUG--UGGCA---UGGCCA--GCGGGCGG ....................((.........(((((....)))))(((((.((((.((((.(((...((........)).))--).)))---).).))--)))))))) ( -30.50) >DroYak_CAF1 4598 105 + 1 GAAGGGGGGCGAAGCGGUGCCCAUAUUCAAAGCGCCAAAUGGCGCGCUCGACUGCAAUGCAACAGCAGCAACAAAAAGUGUGUGUGGCG---UGGCAAGAGCGAGCGG (((...(((((......)))))...)))...(((((....)))))(((((..(((.((((.(((...(((........))).))).)))---).)))....))))).. ( -41.20) >consensus GAAG_GGGGAGAAGCGGUGCCCAUAUUCAAAGCGCCAAAUGGCGCGCUCGACUGCAAUGCAACAGCAGCAACAAAAAGUGUG__UGGCA___UGGCCA__GCGGGCGG ..................((((.........(((((....)))))............(((....)))............((...((((......))))..)))))).. (-19.89 = -21.20 + 1.31)


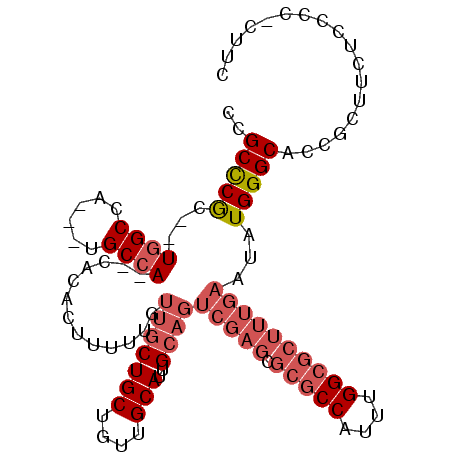
| Location | 10,460,190 – 10,460,281 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -19.98 |
| Energy contribution | -22.60 |
| Covariance contribution | 2.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10460190 91 - 22224390 UCGCCCA---UGGCCAGCGGGCCA--CACACUUUUUGUUGCUGCUGUUGCAUUG------------CCAUUUGGCGCUUUGAAUAUGGGCACCGCCAUUCCCCGCUUC ..(((((---(((((....)))).--.......((((..(((((....)))..(------------((....)))))..)))).))))))...((........))... ( -27.80) >DroSec_CAF1 4660 101 - 1 CCGCCCGC--UGGCCA---UGCCA--CACACUUUUUGUUGCUGCUGUUGCAUUGCAGUCGAGCGCGCCAUUUGGCGCUUUGAAUAUGGGCACCGCUUCUCCCCUCUUC ..(((((.--..((.(---(((.(--(((((........).)).))).)))).))..(((((.(((((....))))))))))...))))).................. ( -31.40) >DroSim_CAF1 4428 101 - 1 CCGCCCGC--UGGCCA---UGCCA--CACACUUUUUGUUGCUGCUGUUGCAUUGCAGUCGAGCGCGCCAUUUGGCGCUUUGAAUAUGGGCNNNNNNNNNNNNNNNNNN ..(((((.--..((.(---(((.(--(((((........).)).))).)))).))..(((((.(((((....))))))))))...))))).................. ( -29.70) >DroYak_CAF1 4598 105 - 1 CCGCUCGCUCUUGCCA---CGCCACACACACUUUUUGUUGCUGCUGUUGCAUUGCAGUCGAGCGCGCCAUUUGGCGCUUUGAAUAUGGGCACCGCUUCGCCCCCCUUC ..((((((((.(((.(---((.((((.(((.....))))).)).))).)))..).)).)))))(((((....))))).........((((........))))...... ( -33.00) >consensus CCGCCCGC__UGGCCA___UGCCA__CACACUUUUUGUUGCUGCUGUUGCAUUGCAGUCGAGCGCGCCAUUUGGCGCUUUGAAUAUGGGCACCGCUUCUCCCC_CUUC ..(((((...((((......)))).............(((((((....)))..))))(((((.(((((....))))))))))...))))).................. (-19.98 = -22.60 + 2.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:27 2006