
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,446,967 – 10,447,110 |
| Length | 143 |
| Max. P | 0.965517 |

| Location | 10,446,967 – 10,447,081 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -20.07 |
| Energy contribution | -21.75 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10446967 114 - 22224390 UUGCGACUAUCUAUGAGUGAUUUCAAUUCCACCAC--CAAUAUU---UUUG-GCGUGGAAUUGGGAACACUCCCCUAAUGCGAUAUAUAAACGAAAGCUGACAACCCUAUUGUACCGCUC ..(((.........(((((.(..(((((((((..(--(((....---.)))-).)))))))))..).)))))......(((((((......(....)..........))))))).))).. ( -33.19) >DroSec_CAF1 5509 110 - 1 UUGCGACAAUCUAUGAGUGAUUUCAAUUCCACCAC------ACU---UUUG-GCGUGGAAUUGGGAACACUCCCCUCAUGCGAUGUGUAAACGAAAGCUGACAACCCUAUUGGACCGCUC ..(((.((((....(((((.(..(((((((((..(------(..---..))-..)))))))))..).)))))...(((.((..(((....)))...))))).......))))...))).. ( -31.50) >DroSim_CAF1 5490 110 - 1 UUGCGACAAUCUAUGAGUGAUUUCAAUUCCACCAC------ACU---UUUG-GCGUGGAAUUGGGAACACUCCCCUCAUGCGAUGUGUAAACGAAAGCUGACAACCCUAUUGGACCGCUC ..(((.((((....(((((.(..(((((((((..(------(..---..))-..)))))))))..).)))))...(((.((..(((....)))...))))).......))))...))).. ( -31.50) >DroEre_CAF1 5780 115 - 1 CUGCUACAU----UUAGUGAUUUCAAUUCCACCACACCAACACCACAUUCG-AUGUGGAAUCGGGAACACUCCCCACAUGCGAUGUGUAACCAAACGCUGACAACCCUAUUGUACCGCUA .(((.((((----(..(((.......................((((((...-))))))....((((....))))..)))..)))))))).......((.(((((.....)))).).)).. ( -20.70) >DroYak_CAF1 6173 105 - 1 UUGCUA-------UUAGUGAUUUCAAUUCCACCAUACCA-CACU--GUUAAAUUGUGGAAUUGGGAACACUCCCCCCAUGCGAUGUGUAAGCAAACGCUGACAACCCUAUUGUG-----A ......-------..((((.(..(((((((((.((....-....--.....)).)))))))))..).))))........(((((((((.(((....))).)))....)))))).-----. ( -24.42) >consensus UUGCGACAAUCUAUGAGUGAUUUCAAUUCCACCAC__CA__ACU___UUUG_GCGUGGAAUUGGGAACACUCCCCUCAUGCGAUGUGUAAACGAAAGCUGACAACCCUAUUGUACCGCUC ..(((.........(((((.(..(((((((((......................)))))))))..).)))))......((((((((((...(....)...)))....))))))).))).. (-20.07 = -21.75 + 1.68)



| Location | 10,447,007 – 10,447,110 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10447007 103 + 22224390 CAUUAGGGGAGUGUUCCCAAUUCCACGCCAAAAAUAUUGGUGGUGGAAUUGAAAUCACUCAUAGAUAGUCGCAAUAUAAUUUGUUCUAUU---------GUGUGUAAUUUAA ........(((((....((((((((((((((.....))))).)))))))))....))))).(((((...(((((((.((....)).))))---------)))....))))). ( -30.70) >DroSec_CAF1 5549 108 + 1 CAUGAGGGGAGUGUUCCCAAUUCCACGCCAAAAGU----GUGGUGGAAUUGAAAUCACUCAUAGAUUGUCGCAAUAUAAUUUAUUCUAUUGUGGGUAUUGUGUGUAAUUCGA ..((((..(((((....(((((((((..((....)----)..)))))))))....))))).........((((((((....((......))...)))))))).....)))). ( -27.00) >DroSim_CAF1 5530 108 + 1 CAUGAGGGGAGUGUUCCCAAUUCCACGCCAAAAGU----GUGGUGGAAUUGAAAUCACUCAUAGAUUGUCGCAAUAUAAUUUAUUCUAUUGUGGGUUUUGUGUGUAAUUCGA ........(((((....(((((((((..((....)----)..)))))))))....)))))...(((((.((((....(((((((......)))))))...)))))))))... ( -27.90) >consensus CAUGAGGGGAGUGUUCCCAAUUCCACGCCAAAAGU____GUGGUGGAAUUGAAAUCACUCAUAGAUUGUCGCAAUAUAAUUUAUUCUAUUGUGGGU_UUGUGUGUAAUUCGA ....(((.(((((....(((((((((((...........)).)))))))))....)))))(((((((((......))))))))))))......................... (-21.45 = -21.57 + 0.11)



| Location | 10,447,007 – 10,447,110 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10447007 103 - 22224390 UUAAAUUACACAC---------AAUAGAACAAAUUAUAUUGCGACUAUCUAUGAGUGAUUUCAAUUCCACCACCAAUAUUUUUGGCGUGGAAUUGGGAACACUCCCCUAAUG ..........(.(---------((((..........))))).).........(((((.(..(((((((((..((((.....)))).)))))))))..).)))))........ ( -28.60) >DroSec_CAF1 5549 108 - 1 UCGAAUUACACACAAUACCCACAAUAGAAUAAAUUAUAUUGCGACAAUCUAUGAGUGAUUUCAAUUCCACCAC----ACUUUUGGCGUGGAAUUGGGAACACUCCCCUCAUG ..((...............(.(((((..........))))).).........(((((.(..(((((((((..(----(....))..)))))))))..).)))))...))... ( -25.00) >DroSim_CAF1 5530 108 - 1 UCGAAUUACACACAAAACCCACAAUAGAAUAAAUUAUAUUGCGACAAUCUAUGAGUGAUUUCAAUUCCACCAC----ACUUUUGGCGUGGAAUUGGGAACACUCCCCUCAUG ..((...............(.(((((..........))))).).........(((((.(..(((((((((..(----(....))..)))))))))..).)))))...))... ( -25.00) >consensus UCGAAUUACACACAA_ACCCACAAUAGAAUAAAUUAUAUUGCGACAAUCUAUGAGUGAUUUCAAUUCCACCAC____ACUUUUGGCGUGGAAUUGGGAACACUCCCCUCAUG .......................(((((...................)))))(((((.(..((((((((((.............).)))))))))..).)))))........ (-21.63 = -21.63 + 0.00)

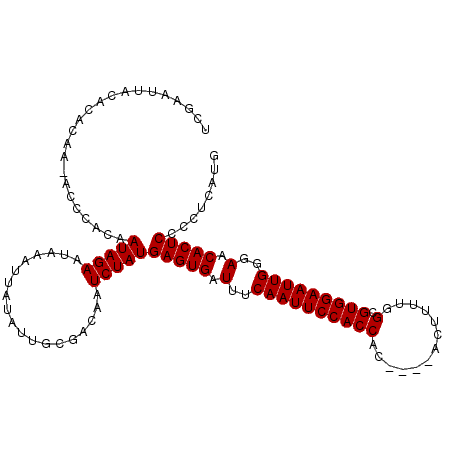
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:18 2006