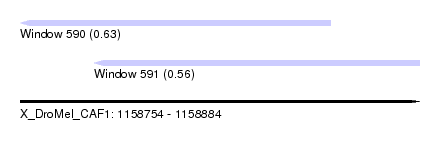
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,158,754 – 1,158,884 |
| Length | 130 |
| Max. P | 0.628720 |

| Location | 1,158,754 – 1,158,855 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1158754 101 - 22224390 CACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCCCCCCCAGUCCAGUUGGCCAAUUAGGCCCAUUUGUCUAUAAUUACCGCCAAUACCA---------------- ((((((((.((.....)).)))).))))....(((...(((((......))))).(((..((((.....))))...)))............))).......---------------- ( -26.50) >DroSec_CAF1 18573 100 - 1 CACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCC-CCCCAGUCCAGUUGGCCAAUUAGGCCCAUUUGUCUAUAAUUACCGCCAAUACCA---------------- ((((((((.((.....)).)))).))))....(((...(((((...-..))))).(((..((((.....))))...)))............))).......---------------- ( -26.30) >DroSim_CAF1 14552 99 - 1 CACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCC--CCCAGUCCAGUUGGCCAAUUAGGCCCAUUUGUCUAUAAUUACCGCCAAUACCA---------------- ((((((((.((.....)).)))).))))....(((...(((((...--.))))).(((..((((.....))))...)))............))).......---------------- ( -26.30) >DroEre_CAF1 23226 108 - 1 CCCAUUGCCAGUUGUUCUGGCAGUCGCGGCGCGGUACC-C-UGCCC--C-----ACAGUUGGCCAAUUAGGCCCAUUUGUCUAUAAUUACCGCCAAUACCAAUAUCUGCCGGUGGGG (((((((((((.....))))))....((((((((((..-.-.....--.-----((((..((((.....))))...)))).......))))))..(((....)))..))))))))). ( -37.49) >consensus CACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCC__CCCAGUCCAGUUGGCCAAUUAGGCCCAUUUGUCUAUAAUUACCGCCAAUACCA________________ ((((((((.((.....)).))))).)))....(((...(((((......))))).(((..((((.....))))...)))............)))....................... (-21.24 = -22.18 + 0.94)

| Location | 1,158,778 – 1,158,884 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -22.06 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1158778 106 - 22224390 UCACUAAUGGUCGGACUUAGCCACCACUUCACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCCCCCCCAGUCCAGUUGGCCAAUUAGGCCCAUU ...(((((((((((.((.((((((.(((.((.((((....))))...)).))).)))))).))..))(((((......)))))......)))).)))))....... ( -30.50) >DroSec_CAF1 18597 105 - 1 UCACUAAUGGUCGGACUUAGCCACCACUUCACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCC-CCCCAGUCCAGUUGGCCAAUUAGGCCCAUU ...(((((((((((.((.((((((.(((.((.((((....))))...)).))).)))))).))..))(((((.(.-.....).))))).)))).)))))....... ( -30.40) >DroSim_CAF1 14576 104 - 1 UCACUAAUGGUCGGACUUAGCCACCACUUCACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCC--CCCAGUCCAGUUGGCCAAUUAGGCCCAUU ...(((((((((((.((.((((((.(((.((.((((....))))...)).))).)))))).))..))((((((..--....).))))).)))).)))))....... ( -30.40) >DroEre_CAF1 23266 94 - 1 UCUCCAAUGGCCGGAGCUA---ACCACUUCCCAUUGCCAGUUGUUCUGGCAGUCGCGGCGCGGUACC-C-UGCCC--C-----ACAGUUGGCCAAUUAGGCCCAUU ........((((.(.((((---((........((((((((.....)))))))).(.((.((((....-)-))).)--)-----.).))))))).....)))).... ( -32.80) >consensus UCACUAAUGGUCGGACUUAGCCACCACUUCACAUUGCCAGUAGUUCUUGCAGUUGUGGCUCGGCACCACUGGCCC__CCCAGUCCAGUUGGCCAAUUAGGCCCAUU .....((((...(((((.((((((.(((.((....((.....))...)).))).)))))).(((.......)))......)))))....((((.....)))))))) (-22.06 = -23.75 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:19 2006