
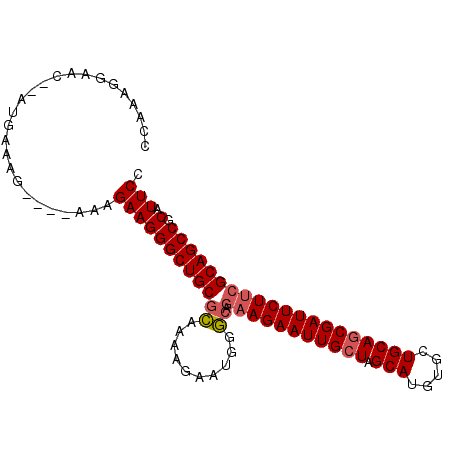
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,390,532 – 10,390,624 |
| Length | 92 |
| Max. P | 0.996221 |

| Location | 10,390,532 – 10,390,624 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.07 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10390532 92 + 22224390 GGAAUGCGGCUGCGAAGAAUCGCUGCAGCACAUGCUAGCAAUUCUUCUGCCCAUUCUUUUGCGCAGCCCUUCUUU----CUUUCAU--GUUCCUUUGG ((((((.(((((((((((((.((((((.....))).))).))))))).((..........)))))))))......----.......--)))))..... ( -29.11) >DroSec_CAF1 34490 92 + 1 GGAAUGCGGCUGCGAAGAAUCGCUGCAGCACAUGCUAGCAAUUCUUCUGCCCAUUCUUUUGCGCAGCCCUUCUUU----CUUUCAU--GUUCCUUUGG ((((((.(((((((((((((.((((((.....))).))).))))))).((..........)))))))))......----.......--)))))..... ( -29.11) >DroEre_CAF1 38838 92 + 1 GGAAUGCGGCUGCGGAGAAUCGCUGCAGCACAUGCUAGCAAUUCUUCUGGCCAUUCUUUGCCGCAGCCCUUCUUU----CUUUCAU--GUUCCUUUAG ((((((.(((((((((((((.((((((.....))).))).))))))).(((........))))))))))......----.......--)))))..... ( -32.81) >DroYak_CAF1 33929 83 + 1 GGAAUGCGGCUGCGAAGAAUCGGUGCAGCACAUGCUAGCAAUUCUU---------------CGCAGCCCUUCUUUCGUUCUUUCAUGUGUUCCUUUGG ((((((((((((((((((((...((((((....))).)))))))))---------------)))))))........(......)..)))))))..... ( -35.50) >consensus GGAAUGCGGCUGCGAAGAAUCGCUGCAGCACAUGCUAGCAAUUCUUCUGCCCAUUCUUUUGCGCAGCCCUUCUUU____CUUUCAU__GUUCCUUUGG ((((.(.(((((((((((((.((((((.....))).))).))))))).((..........)))))))))))))......................... (-24.70 = -25.07 + 0.37)

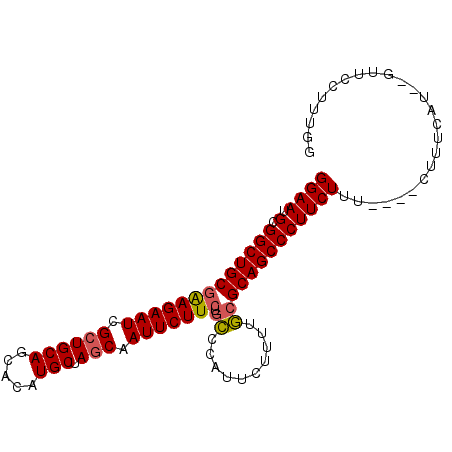

| Location | 10,390,532 – 10,390,624 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10390532 92 - 22224390 CCAAAGGAAC--AUGAAAG----AAAGAAGGGCUGCGCAAAAGAAUGGGCAGAAGAAUUGCUAGCAUGUGCUGCAGCGAUUCUUCGCAGCCGCAUUCC ..........--.......----...((((((((((((..........)).(((((((((((.(((.....)))))))))))))))))))).).))). ( -32.50) >DroSec_CAF1 34490 92 - 1 CCAAAGGAAC--AUGAAAG----AAAGAAGGGCUGCGCAAAAGAAUGGGCAGAAGAAUUGCUAGCAUGUGCUGCAGCGAUUCUUCGCAGCCGCAUUCC ..........--.......----...((((((((((((..........)).(((((((((((.(((.....)))))))))))))))))))).).))). ( -32.50) >DroEre_CAF1 38838 92 - 1 CUAAAGGAAC--AUGAAAG----AAAGAAGGGCUGCGGCAAAGAAUGGCCAGAAGAAUUGCUAGCAUGUGCUGCAGCGAUUCUCCGCAGCCGCAUUCC ..........--.......----...(((((((((((((........))).(.(((((((((.(((.....)))))))))))).))))))).).))). ( -32.30) >DroYak_CAF1 33929 83 - 1 CCAAAGGAACACAUGAAAGAACGAAAGAAGGGCUGCG---------------AAGAAUUGCUAGCAUGUGCUGCACCGAUUCUUCGCAGCCGCAUUCC .....((((..(.........(....)...(((((((---------------((((((((.((((....))))...))))))))))))))))..)))) ( -34.00) >consensus CCAAAGGAAC__AUGAAAG____AAAGAAGGGCUGCGCAAAAGAAUGGGCAGAAGAAUUGCUAGCAUGUGCUGCAGCGAUUCUUCGCAGCCGCAUUCC ..........................((((((((((((..........)).(((((((((((.(((.....)))))))))))))))))))).).))). (-26.69 = -27.50 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:04 2006