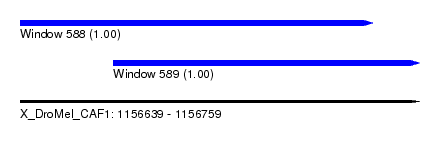
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,156,639 – 1,156,759 |
| Length | 120 |
| Max. P | 0.999965 |

| Location | 1,156,639 – 1,156,745 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.42 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -21.40 |
| Energy contribution | -24.52 |
| Covariance contribution | 3.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.97 |
| SVM RNA-class probability | 0.999965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
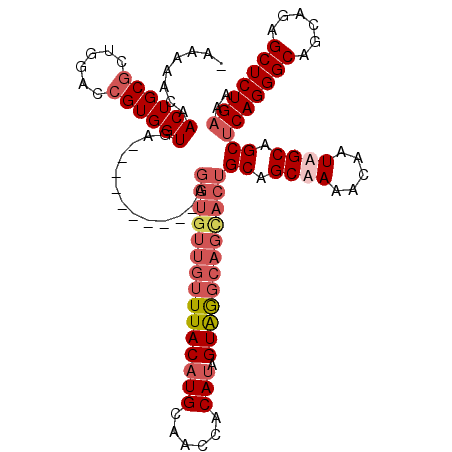
>X_DroMel_CAF1 1156639 106 + 22224390 -AAAUACAACUGCGCUGGACCGUGGUGA----------AGGUGUUGUUUACAUGCAACUACAUAGUAGGCAGCACUGCAGCUAAACAAUAGCAGCUCAGGGCAGCAAAGCUCUGAAA -..........((((((.....((((..----------.(((((((((((((((......))).))))))))))))...)))).....)))).))(((((((......))))))).. ( -38.40) >DroSec_CAF1 16414 106 + 1 -NNNNNNNNNNNNNNNNNNNNNNNNNNN----------NNNNNNNNNUUACAUGCAACUACAUAGUAGGCAGCACUGCUGCUAAACAAUAGCGGCCCAGGGCAGCAGAGCUCUGAAA -...........................----------..............(((..((((...))))...)))..(((((((.....))))))).((((((......))))))... ( -22.40) >DroEre_CAF1 16837 115 + 1 -AAAAACAACUGCACUGGACCGUGGUGAAGGUGUUGUAAGGUGUUGUUUACAUGCAA-CACAUAGUAGGCAGUACUGCAGCAAAACAAUAGCAGCUCAGGGCAGCAGAGCUCUGAAA -........((((.((..((....))..)).((((((...(((((((......))))-)))...((((......))))......)))))))))).(((((((......))))))).. ( -35.80) >DroYak_CAF1 16828 117 + 1 GAAAAACAACUGCGCUGGACCGUGGUGAAGGUGUUGUAAGGUGUUGUUUACAUGCAACCACACAGUGAGCAGCACUGCAGCAAAACAAUAGCAGCUCAGGGCAGCAGAGCUCUGAAA .........((((.((..((....))..)).((((((..((((((((((((.((........))))))))))))))))))))........)))).(((((((......))))))).. ( -39.50) >consensus _AAAAACAACUGCGCUGGACCGUGGUGA__________AGGUGUUGUUUACAUGCAACCACAUAGUAGGCAGCACUGCAGCAAAACAAUAGCAGCUCAGGGCAGCAGAGCUCUGAAA ........((((((......)))))).............(((((((((((((((......))).))))))))))))((.((((.....)))).))(((((((......))))))).. (-21.40 = -24.52 + 3.13)

| Location | 1,156,667 – 1,156,759 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -9.38 |
| Energy contribution | -13.58 |
| Covariance contribution | 4.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
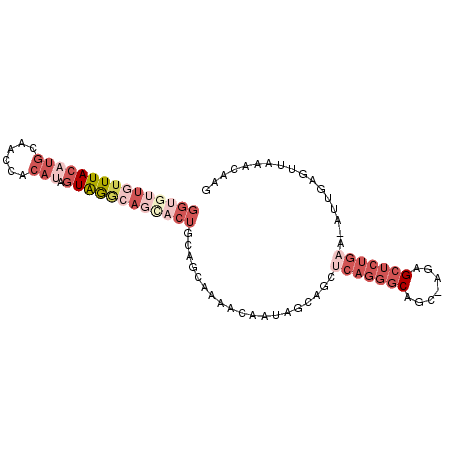
>X_DroMel_CAF1 1156667 92 + 22224390 GGUGUUGUUUACAUGCAACUACAUAGUAGGCAGCACUGCAGCUAAACAAUAGCAGCUCAGGGCAGC-AAAGCUCUGAA-AUUGAGUUAAACAAG (((((((((((((((......))).))))))))))))((.((((.....)))).))(((((((...-...))))))).-............... ( -31.20) >DroSec_CAF1 16442 92 + 1 NNNNNNNNUUACAUGCAACUACAUAGUAGGCAGCACUGCUGCUAAACAAUAGCGGCCCAGGGCAGC-AGAGCUCUGAA-AUUGAGUUAAACAAG .............(((..((((...))))...)))..(((((((.....))))))).((((((...-...))))))..-............... ( -22.40) >DroEre_CAF1 16875 91 + 1 GGUGUUGUUUACAUGCAA-CACAUAGUAGGCAGUACUGCAGCAAAACAAUAGCAGCUCAGGGCAGC-AGAGCUCUGAA-AAUGACUAAAACAAU .(((((((......))))-)))...((((......))))..........(((((..(((((((...-...))))))).-..)).)))....... ( -26.40) >DroYak_CAF1 16867 92 + 1 GGUGUUGUUUACAUGCAACCACACAGUGAGCAGCACUGCAGCAAAACAAUAGCAGCUCAGGGCAGC-AGAGCUCUGAA-ACUCAGUUAAACAAU ((((((((((((.((........))))))))))))))((.((.........)).))(((((((...-...))))))).-............... ( -27.40) >DroAna_CAF1 9904 78 + 1 GGUAUUGUUUACAACCAACAACA-ACUAAAAA---CUA-AAAAAAACAAUA-----------CAACAAAAGCUUUGAAAAUUGAGUUAAACAAC .(((((((((.............-........---...-....))))))))-----------)......(((((........)))))....... ( -8.39) >consensus GGUGUUGUUUACAUGCAACCACAUAGUAGGCAGCACUGCAGCAAAACAAUAGCAGCUCAGGGCAGC_AGAGCUCUGAA_AUUGAGUUAAACAAG (((((((((((((((......))).))))))))))))...................(((((((.......)))))))................. ( -9.38 = -13.58 + 4.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:17 2006