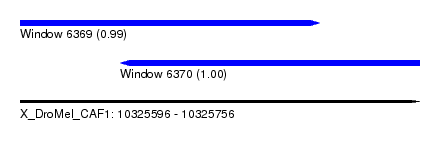
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,325,596 – 10,325,756 |
| Length | 160 |
| Max. P | 0.995417 |

| Location | 10,325,596 – 10,325,716 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -29.90 |
| Energy contribution | -31.40 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10325596 120 + 22224390 AACCAGGGUCAAAAAUGAAAAGGAUGAGAAGAAUGACAACGAUGAGACUGAUUCGCCGCAUGAGGCUGCCGAGAUCGAGGAGCCCGAUGAAGAGGUGGAUGAGGCUGAUCAGGAGACCAA ......((((.....(....)...................(((.((.((.(((((((.(((..((((.(((....)).).))))..)))....))))))).)).)).)))....)))).. ( -35.00) >DroSec_CAF1 3768 114 + 1 ------GGUAAAAAAUGAAAAGGAUGAGAAGAGUGACAACGAUGAGACUGAUUCGCCGCAUGAGGCUGCCGAGAUCGAGGAGCCCGAUGAAGAGGUGGAUGAGGCUGAUCAAGGGACCAA ------(((.......................((....))(((.((.((.(((((((.(((..((((.(((....)).).))))..)))....))))))).)).)).))).....))).. ( -33.30) >DroSim_CAF1 3530 114 + 1 ------GGUCAAAAAUGAAAAGGAUGAGAAGAGUGACAACGAUGAGACUGAUUCGCCGCAUGAGGCUGCCGAGAUCGAGGAGCCCGAUGAAGAGGUGGAUGAGGCUGAUCAAGGGACCAA ------((((......................((....))(((.((.((.(((((((.(((..((((.(((....)).).))))..)))....))))))).)).)).)))....)))).. ( -36.80) >DroEre_CAF1 4043 120 + 1 AGACAAGGAGAAAAACGAUAAGGAGGAUAAGAGUGACAACGAUGAGACUGAUUCGCCGCAGGAGGCUGCCGAGAUCGAGGAGCCCGAUGAGGAGGUGGAUGAUGCUGAUCAGGAGGCCAA ........................((......((....))(((.((..(.(((((((.((...((((.(((....)).).))))...))....))))))).)..)).)))......)).. ( -27.50) >consensus ______GGUCAAAAAUGAAAAGGAUGAGAAGAGUGACAACGAUGAGACUGAUUCGCCGCAUGAGGCUGCCGAGAUCGAGGAGCCCGAUGAAGAGGUGGAUGAGGCUGAUCAAGAGACCAA ......((((......................((....))(((.((.((.(((((((.(((..((((.(((....)).).))))..)))....))))))).)).)).)))....)))).. (-29.90 = -31.40 + 1.50)



| Location | 10,325,636 – 10,325,756 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -39.29 |
| Energy contribution | -39.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10325636 120 - 22224390 GCCCUCCUUCUGACUGCCGUAAGCAGGCUGCUUGGUGGUCUUGGUCUCCUGAUCAGCCUCAUCCACCUCUUCAUCGGGCUCCUCGAUCUCGGCAGCCUCAUGCGGCGAAUCAGUCUCAUC .........((((.(((((((.(.((((((((.(((((..((((((....))))))......))))).....((((((...))))))...))))))))).)))))))..))))....... ( -44.70) >DroSec_CAF1 3802 120 - 1 ACCCUCCUUCUGACUGUCGUAAACAGGCUGCUUGGUGGUCUUGGUCCCUUGAUCAGCCUCAUCCACCUCUUCAUCGGGCUCCUCGAUCUCGGCAGCCUCAUGCGGCGAAUCAGUCUCAUC .........((((.(((((((...((((((((.(((((..((((((....))))))......))))).....((((((...))))))...))))))))..)))))))..))))....... ( -39.60) >DroSim_CAF1 3564 120 - 1 ACCCUCCUUCUGACUGCCGUAAACAGGCUGCUUGGUGGUCUUGGUCCCUUGAUCAGCCUCAUCCACCUCUUCAUCGGGCUCCUCGAUCUCGGCAGCCUCAUGCGGCGAAUCAGUCUCAUC .........((((.(((((((...((((((((.(((((..((((((....))))))......))))).....((((((...))))))...))))))))..)))))))..))))....... ( -42.30) >DroEre_CAF1 4083 120 - 1 ACCCUCUUUCUUACUGCCGUUAACAGGCUGCUUGGUGGUCUUGGCCUCCUGAUCAGCAUCAUCCACCUCCUCAUCGGGCUCCUCGAUCUCGGCAGCCUCCUGCGGCGAAUCAGUCUCAUC ..............((((((....((((((((.(((((.....((..........)).....))))).....((((((...))))))...))))))))...))))))............. ( -36.80) >consensus ACCCUCCUUCUGACUGCCGUAAACAGGCUGCUUGGUGGUCUUGGUCCCCUGAUCAGCCUCAUCCACCUCUUCAUCGGGCUCCUCGAUCUCGGCAGCCUCAUGCGGCGAAUCAGUCUCAUC .........((((.(((((((...((((((((.(((((..((((((....))))))......))))).....((((((...))))))...))))))))..)))))))..))))....... (-39.29 = -39.85 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:48 2006