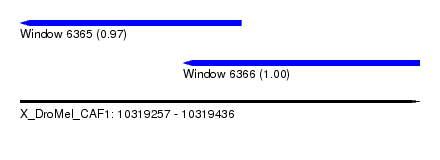
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,319,257 – 10,319,436 |
| Length | 179 |
| Max. P | 0.999796 |

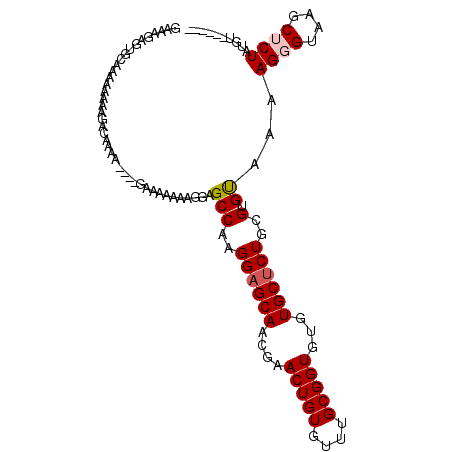
| Location | 10,319,257 – 10,319,356 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10319257 99 - 22224390 GAAAGAGUGCAAAAAAAAGGCGAAG----GAAAGAAAGAAGCCAAGGAGCAACGAACUGUGUUUGCGGUGUGUGCUCUGCGUGCGAAAGGGUAAGCUCUAUGG------ ...(((((((........(((....----...........)))..((((((.((.(((((....))))).))))))))))...(....).....)))))....------ ( -26.66) >DroVir_CAF1 63009 103 - 1 AGAAGAGUGCAAAAAACGUACAAAAGAAAGAAAAAAAGGAGCCAAGGAGCAACGAACUGUGUGUGCGGUGUAUGCUCUGAGUGUAAAAGGGUAAGCACUAUAU------ .....(((((...(..(.((((...............(....)..((((((....(((((....)))))...))))))...))))...)..)..)))))....------ ( -20.90) >DroPse_CAF1 60506 89 - 1 GAAAGAGUGCAAAAAA--------------------AGGAGCCAAGGAGCAACGAACUGUGUUUGCGGUGUGUGCGCUGCGUGUAAAAGGGUAAGCUCUAUGUGCCAAC ................--------------------..(.((((.(((((......((.((..(((((((....)))))))..))...))....))))).)).)))... ( -21.00) >DroYak_CAF1 47853 99 - 1 GAAAGAGUGCAAAAAAAAGGCGAAG----GAAAGAAAGAAGCCAAGGAGCAACGAACUGUGUUUGCGGUGUGUGCUCUGCGUGCGAAAGGGUAAGCUCUAUGA------ ...(((((((........(((....----...........)))..((((((.((.(((((....))))).))))))))))...(....).....)))))....------ ( -26.66) >DroMoj_CAF1 75275 103 - 1 AGAAGAGUGCAAAAAACGUACAAAAAGAAAGAAAAAAGGAGCCAAGGAGCAACGAACUGUGUGUGCGGUGUAUGCUCUGAGUGUAAAAGGGUAAGCACUAUAU------ .....(((((...(..(.((((...............(....)..((((((....(((((....)))))...))))))...))))...)..)..)))))....------ ( -20.90) >consensus GAAAGAGUGCAAAAAAAAGACAAAA____GAAAAAAAGGAGCCAAGGAGCAACGAACUGUGUUUGCGGUGUGUGCUCUGCGUGUAAAAGGGUAAGCUCUAUGU______ ........................................(((..((((((....(((((....)))))...))))))..).))...((((....)))).......... (-15.40 = -15.76 + 0.36)

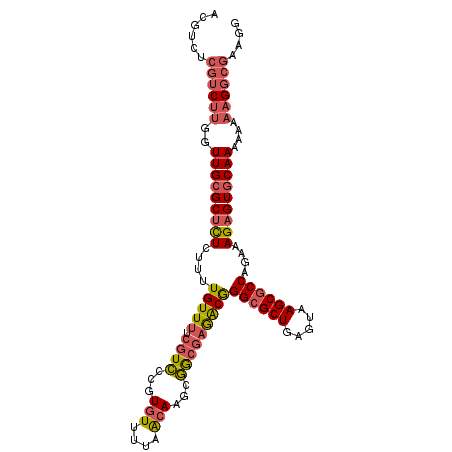
| Location | 10,319,330 – 10,319,436 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -25.05 |
| Energy contribution | -27.37 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10319330 106 - 22224390 ACGUCUCGUCUUGGUUGCGCUCUCUUUUGUUUUCGUCCCGUGUUUUAACAAGCGGCGAAACGGGCGCUGAGUAAGCGCCAGAAAGAGUGCAAAAAAAAGGCGAAGG .....(((((((..(((((((((.((((..(((((((.(.(((....))).).)))))))..((((((.....)))))))))))))))))))....)))))))... ( -43.00) >DroPse_CAF1 60579 92 - 1 GCAUUUCGU-UUCGUUGCGCAUUUUGUUGUUUUCAUU---UUUUUGUGCAACCAACACGGCAGGGGCUGAGUAAGCGCCAGAAAGAGUGCAAAAAA---------- ((((((..(-(((((((((((......((....))..---....))))))))..........((.(((.....))).)).))))))))))......---------- ( -27.40) >DroYak_CAF1 47926 106 - 1 ACGUCACGUCUUGGUUGAGCUCUCUUUUGUUUUCGUCCCGUGUUUUCACAAGCGGCGAGACGGGCGCUGAGUAAGCGCCAGAAAGAGUGCAAAAAAAAGGCGAAGG ......((((((..((..((.(((((((.....((((.(((((((....)))).))).))))((((((.....)))))).))))))).))..))..)))))).... ( -40.90) >consensus ACGUCUCGUCUUGGUUGCGCUCUCUUUUGUUUUCGUCCCGUGUUUUAACAAGCGGCGAGACGGGCGCUGAGUAAGCGCCAGAAAGAGUGCAAAAAAAAGGCGAAGG ......((((((..(((((((((....(((((.((((...(((....)))...)))))))))((((((.....))))))....)))))))))....)))))).... (-25.05 = -27.37 + 2.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:44 2006