| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,311,627 – 10,311,726 |
| Length | 99 |
| Max. P | 0.878304 |

| Location | 10,311,627 – 10,311,726 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -55.23 |
| Consensus MFE | -21.14 |
| Energy contribution | -23.33 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
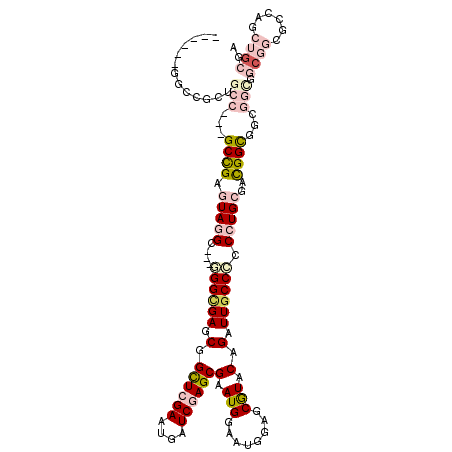
>X_DroMel_CAF1 10311627 99 + 22224390 ------GGCCGCCGCC---GCCGAGUAGGC---GGGCGAACGGCUCGAAUGAUCGAGCGAAUGGAAUGGAGCGUACAGAUUACCCCCCUGCGACGGU------GGC---GGCAUCUGCGA ------....((((((---((((.(((((.---(((..(.(.((((((....))))))(.(((........))).).).)..))).)))))..))))------)))---)))........ ( -46.90) >DroVir_CAF1 55026 108 + 1 GGCAGCAGCUGCUGCC---GCUGAGUAGGC---GGGCGAGCGGUUCGAAUGAUCGAGCGAAUGGAAUGGAGCGUACAGAUUGCCCGCCUGCGAUGGCGG------CGGCGCCAGCUGCGA .(((((.((.((((((---(((..((((((---((((((.(.((((((....))))))(.(((........))).).).))))))))))))...)))))------))))))..))))).. ( -62.00) >DroGri_CAF1 49937 114 + 1 GGCAGCGGCCGCUGCC---GCUGAAUAGGC---GGGUGAGCGGCUCGAAUGAUCAAGCGAAUGGAAUGGAGCGUACAGAUUGCCCGCCUGCGAUGGUGGCGGCGGCGGCGCCAGCUGCGA (((.((.(((((((((---((((..(((((---(((..((((.(((..((..(((......))).)).)))))).....)..))))))))...))))))))))))).)))))........ ( -58.70) >DroMoj_CAF1 67134 108 + 1 GGCGGC------GGCC---GCUGAGUAUGC---GGGCGAGCGGUUCGAAUGAUCAAGCGAAUGGAAUGAAGCGUACAGAUUGCCAGCCUGCGAUGGCGGCGGCGGCGGCGCCAGCUGCGA .(((((------.(((---((((....(((---((((..(((((..(.(((.(((..(.....)..)))..))).)..)))))..)))))))....)))))))(((...))).))))).. ( -49.10) >DroAna_CAF1 50552 108 + 1 ------GGCCGCCGCCGCUGCCGAGUAUGCAGCUGGCGAGCGGCUCGAAUGAUCGAGCGAAUGGAAUGGAGCGUACAGAUUGCCGCCCUGCGACGGCGGCGGUG------GCAUCUGCGA ------....(((((((((((((....(((((..((((.(((((((((....))))))(.(((........))).)....)))))))))))).)))))))))))------))........ ( -54.40) >DroPer_CAF1 54144 108 + 1 ------GGCCGCUGCC---GCCGAAUAGGC---GGGCGAGCGGCUCGAAUGAUCGAGCGUAUGGAAUGGAGCAUACAGAUUGCCGCCCUGCGACGGUGGCGGCGGCGGUGGCAUCUGCGA ------.(((((((((---((((.....((---(((((.(((((((((....))))))(((((........)))))....)))))))).)).....))))))))))))).((....)).. ( -60.30) >consensus ______GGCCGCUGCC___GCCGAGUAGGC___GGGCGAGCGGCUCGAAUGAUCGAGCGAAUGGAAUGGAGCGUACAGAUUGCCCCCCUGCGACGGCGGCGGCGGCGGCGCCAGCUGCGA .............(((...((((.(((((....((((((.(.((((((....))))))(.(((........))).).).)))))).)))))..))))...))).((((......)))).. (-21.14 = -23.33 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:41 2006