| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,148,517 – 1,148,622 |
| Length | 105 |
| Max. P | 0.586091 |

| Location | 1,148,517 – 1,148,622 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -25.79 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1148517 105 + 22224390 AGCUCGCUCCUCCGCCGAGGGCGUAAACUUGAGCAGCUGCUCGACCAUGUCCAGCUGCAGCUGCUCGUUGCUGUCCAUGGAAAGAAUGGCCCUGAA-AGCGGCA----------U-U .((.((((..((.((((.((((((((...(((((((((((..(((...))).....))))))))))))))).)))).(....)...))))...)).-)))))).----------.-. ( -39.60) >DroGri_CAF1 43656 109 + 1 AGCACGCUCCUCCGCCGAGGGCGUAAACUUCAGCAGCUGCUCAACCAUGUCCAGUGCCAGCUGCUCAUUGCUAUCCAUCGAGAGUAUAGCUCUAAG-AAACACAACACA-------A (((((((.((((....)))))))).......((((((((....((........))..))))))))....)))........((((.....))))...-............-------. ( -29.90) >DroEre_CAF1 8532 105 + 1 AGCUCGCUCCUCCGCCGAGGGCGUGAACUUGAGCAGUUGCUCGACCAUGUCCAGUUGCAGCUGCUCGUUGCUGUCCAUGGAGAGGAUGGCCCUGAA-AGUGGUA----------U-G .((.((.(((((..(((.(((((..(...(((((((((((..(((...))).....))))))))))))..).)))).))).)))))))))((....-...))..----------.-. ( -39.00) >DroYak_CAF1 8505 106 + 1 AGCUCGCUCCUCCGCAGAGGGCGUGAACUUGAGCAGCUGCUCGACCAUGUCCAGUUGCAGCUGCUCGUUGCUGUCCAUGGAGAGAAUGGCUCUGAA-AGUGGCA----------UUU .((.((((.(((((..((.((((......(((((((((((..(((...))).....))))))))))).)))).))..)))))(((.....)))...-)))))).----------... ( -40.40) >DroMoj_CAF1 56443 112 + 1 CGCACGCUCCUCGGCAGAUGGUGUAAACUUGAGCAGCUGCUCGACCAUGUCCAGCGCCAGCUGCUCAUUGCUAUCCAUGGAGAGUAUGGCUCUGUA-AGGGA----AAUUAGACACC .((.((.....))))....(((((.....((((((((((((.(((...))).)))...)))))))))......(((.((.((((.....)))).))-.))).----......))))) ( -36.50) >DroAna_CAF1 1545 114 + 1 GGCGCGCUCCUCCGCCGAUGGUGUGAACUUCAGUAGCUGUUCCACCAUGUCCAGAGCCAGCUGCUCGUUGCUGUCCAUGGAGAGAAUGGCUCUGAGGAGAUGCA---CAAAAGGUUU .(.(((((((((.((((((((((.((((..(....)..)))))))))).((((..(.((((........)))).)..)))).....))))...)))))).))).---)......... ( -42.50) >consensus AGCUCGCUCCUCCGCCGAGGGCGUAAACUUGAGCAGCUGCUCGACCAUGUCCAGUGCCAGCUGCUCGUUGCUGUCCAUGGAGAGAAUGGCUCUGAA_AGAGGCA__________U_U .((.((.(((((((..((.((((......((((((((((((.(((...))).))...)))))))))).)))).))..))))).)).))))........................... (-25.79 = -26.52 + 0.73)

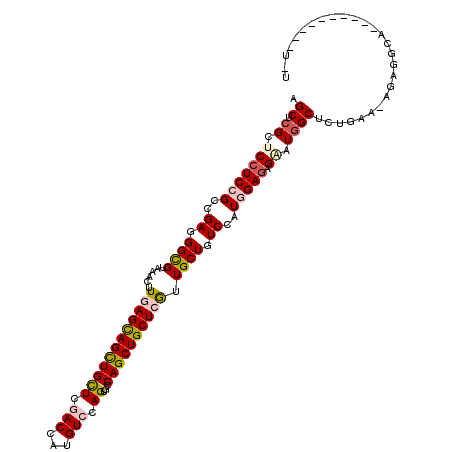
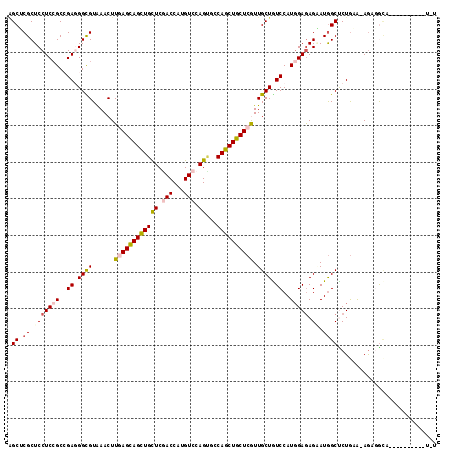
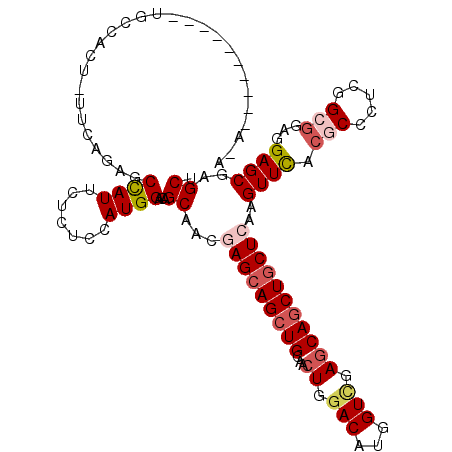
| Location | 1,148,517 – 1,148,622 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -24.53 |
| Energy contribution | -25.53 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1148517 105 - 22224390 A-A----------UGCCGCU-UUCAGGGCCAUUCUUUCCAUGGACAGCAACGAGCAGCUGCAGCUGGACAUGGUCGAGCAGCUGCUCAAGUUUACGCCCUCGGCGGAGGAGCGAGCU .-.----------.((.(((-(((((((........))).)))).)))...((((((((((.(((......)))...)))))))))).......(((((((....)))).))).)). ( -40.10) >DroGri_CAF1 43656 109 - 1 U-------UGUGUUGUGUUU-CUUAGAGCUAUACUCUCGAUGGAUAGCAAUGAGCAGCUGGCACUGGACAUGGUUGAGCAGCUGCUGAAGUUUACGCCCUCGGCGGAGGAGCGUGCU .-------..((((((.(.(-(..((((.....)))).)).).)))))).(.((((((((.((((......)).))..)))))))).)....(((((((((....)))).))))).. ( -36.20) >DroEre_CAF1 8532 105 - 1 C-A----------UACCACU-UUCAGGGCCAUCCUCUCCAUGGACAGCAACGAGCAGCUGCAACUGGACAUGGUCGAGCAACUGCUCAAGUUCACGCCCUCGGCGGAGGAGCGAGCU .-.----------.......-.....((((((....((((..(.((((........)))))...)))).))))))((((....)))).(((((..((((((....)))).))))))) ( -33.90) >DroYak_CAF1 8505 106 - 1 AAA----------UGCCACU-UUCAGAGCCAUUCUCUCCAUGGACAGCAACGAGCAGCUGCAACUGGACAUGGUCGAGCAGCUGCUCAAGUUCACGCCCUCUGCGGAGGAGCGAGCU ...----------(((((..-...((((.....))))...))).))((...((((((((((.(((......)))...)))))))))).......(((((((....)))).))).)). ( -38.80) >DroMoj_CAF1 56443 112 - 1 GGUGUCUAAUU----UCCCU-UACAGAGCCAUACUCUCCAUGGAUAGCAAUGAGCAGCUGGCGCUGGACAUGGUCGAGCAGCUGCUCAAGUUUACACCAUCUGCCGAGGAGCGUGCG (((((..((((----..((.-...((((.....))))....)).......(((((((((...(((.(((...))).)))))))))))))))).)))))....((((.....)).)). ( -36.10) >DroAna_CAF1 1545 114 - 1 AAACCUUUUG---UGCAUCUCCUCAGAGCCAUUCUCUCCAUGGACAGCAACGAGCAGCUGGCUCUGGACAUGGUGGAACAGCUACUGAAGUUCACACCAUCGGCGGAGGAGCGCGCC .........(---(((..((((((...(((......((((.((.((((........)))).)).)))).((((((((((..(....)..)))).)))))).))).)))))).)))). ( -44.90) >consensus A_A__________UGCCACU_UUCAGAGCCAUUCUCUCCAUGGACAGCAACGAGCAGCUGCAACUGGACAUGGUCGAGCAGCUGCUCAAGUUCACGCCCUCGGCGGAGGAGCGAGCU ............................((((.......))))...((...(((((((((...((.(((...))).)))))))))))..((((.(((.....)))...))))..)). (-24.53 = -25.53 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:14 2006