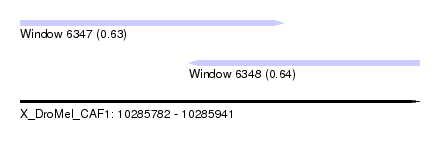
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,285,782 – 10,285,941 |
| Length | 159 |
| Max. P | 0.639844 |

| Location | 10,285,782 – 10,285,887 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.28 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -14.08 |
| Energy contribution | -16.20 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
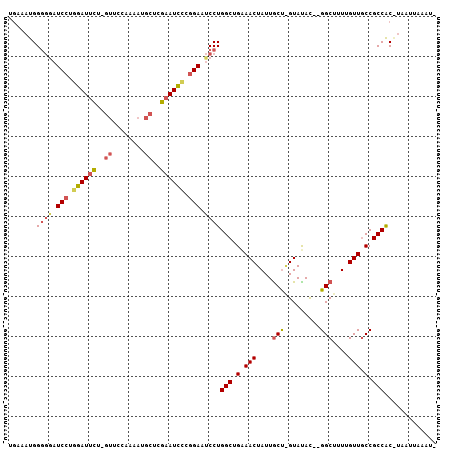
>X_DroMel_CAF1 10285782 105 + 22224390 UGAAAUGGGCGAUCCUAGAUUCUUGUUCCAAAAUGCUCGAAUUCCGGAAACCUGGCUGGAACUAUUACUAGUAUAC--GGCUUUUGUUGCCGCCACGUAAUUAAAUA ....(((((((.(((..(((((..((........))..)))))..)))(((..(((((..((((....))))...)--))))...)))..)))).)))......... ( -25.00) >DroSec_CAF1 25524 94 + 1 UGAAAUGGGGGAUCCUGGAUUCUGGUUCCAAAAUGCUCGAAUCCCGGAAUCCUGGC-GAAACUAUUGCC-GUAUACUAGGCUUUUGUUACCGCCAC----------- .......((((.(((.((((((.(((........))).)))))).))).))))(((-(.(((....(((-........)))....)))..))))..----------- ( -33.20) >DroYak_CAF1 14091 99 + 1 UCAAAUGGCGAAUCCAGGAUCUU------AGAAUCUUAGAAUCUUAGAUUCCUGGCUGAAACUAUUGCUGGUACAU--GGCUUUGGUUGCCGCCGCAUAAUUAAAUG ......((((...(((((((((.------(((.((...)).))).))).))))))....(((((..((((.....)--)))..)))))..))))............. ( -27.90) >consensus UGAAAUGGGGGAUCCUGGAUUCU_GUUCCAAAAUGCUCGAAUCCCGGAAUCCUGGCUGAAACUAUUGCU_GUAUAC__GGCUUUUGUUGCCGCCAC_UAAUUAAAU_ .......((((.(((.((((((..((........))..)))))).))).))))(((.(.(((....(((.........)))....))).).)))............. (-14.08 = -16.20 + 2.12)


| Location | 10,285,849 – 10,285,941 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -16.12 |
| Consensus MFE | -10.37 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10285849 92 - 22224390 CCAUUAUUAUUUCCCAUGCUCUCUGACACGGC--ACUCAUAAUUAAACGCAUUUUGUAUUUAAUUACGUGGCGGCAACAAAAGCC--GUAUACUAG ...........................(((((--.(.(.((((((((..(.....)..)))))))).).)..(....)....)))--))....... ( -16.20) >DroPse_CAF1 14508 81 - 1 CC--CAUCAUUUCCCCGGAGCUCUGACACGGU--GCUCAUAAUUAAACGCAUUUUGUAUUUAAUUAUGAGACGACGAAAACAGCC----------- ..--.....((((..(((....)))...((..--.((((((((((((..(.....)..)))))))))))).))..))))......----------- ( -18.70) >DroEre_CAF1 13710 80 - 1 CCAUUAUUAUUUCCCAUGGGCUCUGACACGGU--ACUCAUAAUUAAACGCAUUUUGUAUUUAAUUAUGUGGCAGCAACCAAA-------------- ................(((...(((.......--.(.((((((((((..(.....)..)))))))))).).)))...)))..-------------- ( -14.40) >DroWil_CAF1 25393 86 - 1 CC--CAUUAUUUCCCCUGGGUCCUGACACGGCACACUCAUAAUUAAGCGCAUUUUGUAUUUAAUUAUGUGACAGCAAAAAAAAAAAAA-------- ((--((..........))))..........((...(.((((((((((..(.....)..)))))))))).)...)).............-------- ( -14.20) >DroYak_CAF1 14152 92 - 1 CCAUUAUUAUUUCCCAUGGGCUCUGACACGGC--ACUCAUAAUUAAACGCAUUUUGCAUUUAAUUAUGCGGCGGCAACCAAAGCC--AUGUACCAG ..............(((((.((.((.....((--.(.((((((((((.((.....)).)))))))))).)))(....))).))))--)))...... ( -22.40) >DroAna_CAF1 16286 92 - 1 CC--CAUUAUUUCCCAAAGGCUCUGACACGGC--ACUCAUAAUUAAACGCAUUUUCUAUUUAAUUAUGUGACAGCAGCAACAGCAACACAUAACGG .(--(.((((.........((((((.((((..--...)(((((((((...........)))))))))))).))).)))...........)))).)) ( -10.85) >consensus CC__CAUUAUUUCCCAUGGGCUCUGACACGGC__ACUCAUAAUUAAACGCAUUUUGUAUUUAAUUAUGUGACAGCAACAAAAGCC__A________ ..............................((...(.((((((((((.((.....)).)))))))))).)...))..................... (-10.37 = -10.07 + -0.30)

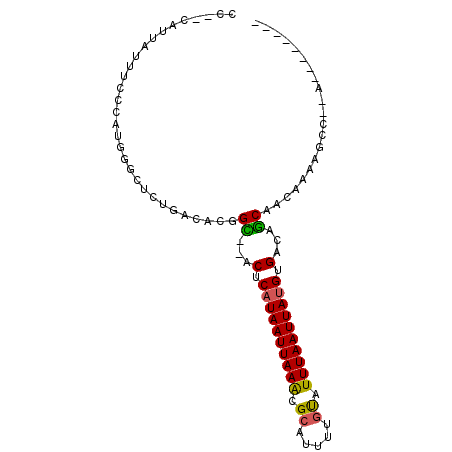

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:27 2006