| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,264,172 – 10,264,264 |
| Length | 92 |
| Max. P | 0.733622 |

| Location | 10,264,172 – 10,264,264 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.09 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -13.98 |
| Energy contribution | -13.73 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
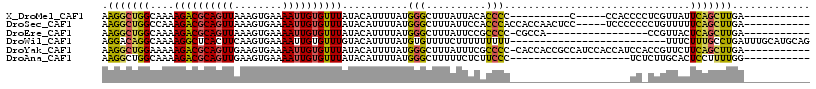
>X_DroMel_CAF1 10264172 92 - 22224390 AAGGCUGGCAAAAGACGCAGUUAAAGUGAAAAUUGUGUUUAUACAUUUUAUGGGCUUUAUUACACCCC----------C-----CCACCCCUCGUUAUUCAGCUUGA----------- .(((((((....((((((((((........))))))))))..........((((..............----------)-----)))...........)))))))..----------- ( -19.14) >DroSec_CAF1 29808 102 - 1 AAGGCUGGCCAAAGACGCAGUUAAAGUGAAAAUUGUGUUUAUACAUUUUAUGGGCUUUAUUCCACCCACCACCAACUCC-----UCCCCCCCUGUUUUUCAGCUUGA----------- .(((((((....((((((((((........))))))))))..........((((..........))))...........-----..............)))))))..----------- ( -19.90) >DroEre_CAF1 37004 89 - 1 AAGGCUGGCAAAAGACGCAGUUAAAGUGAAAAUUGUGUUUAUACAUUUUAUGGGCUUUAUUCCGCCCC-CGCCA-----------------CCGUUACUCAGCUUGA----------- ..((.((((...((((((((((........))))))))))...........((((........)))).-.))))-----------------))..............----------- ( -24.80) >DroWil_CAF1 35863 92 - 1 AGGACAGGCAAAAGGCUCACUUCAAGUGAAAAUUGUGUUUGUACAUUUUAUGUGUUUUCUUUUUUUUU--------------------------UUUCUUUGCCUGAUUUGCAUGCAG ....((((((((..(.((((.....))))(((.((((....)))).)))...................--------------------------.)..))))))))............ ( -16.60) >DroYak_CAF1 30174 106 - 1 AAGGCUGGAAAAAGACGCAGUUGAAGUGAAAAUUGUGUUUAUACAUUUUAUGGGCUUUAUUUCGCCCC-CACCACCGCCAUCCACCAUCCACCGUUCUUCAGCUUGA----------- .((((((((...((((((((((........))))))))))...........((((........)))).-............................))))))))..----------- ( -25.30) >DroAna_CAF1 36234 87 - 1 AAGGCUGGCAAAAGACGCAGUUGAAGUGAAAAUUGUGUUUAUACAUUUUAUGGGCUUUUUCUCUUCCC--------------------UCUCUUGCACUCCUUUUGG----------- ((((...((((.(((.(.....((((.(((((....(((((((.....))))))).))))).))))).--------------------))).))))...))))....----------- ( -18.80) >consensus AAGGCUGGCAAAAGACGCAGUUAAAGUGAAAAUUGUGUUUAUACAUUUUAUGGGCUUUAUUUCACCCC____________________CC_CCGUUACUCAGCUUGA___________ .(((((((....((((((((((........))))))))))...........(((..........)))...............................)))))))............. (-13.98 = -13.73 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:25 2006