
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,233,779 – 10,233,970 |
| Length | 191 |
| Max. P | 0.999843 |

| Location | 10,233,779 – 10,233,870 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 62.85 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.48 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
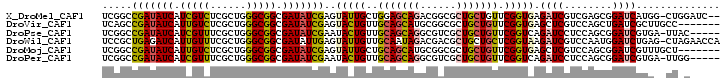
>X_DroMel_CAF1 10233779 91 + 22224390 AAUUACUCGUCGAUGGCGGCGGUGGCCUCCUUUGGAUC------CGGAUACGGAUCGGGAUCUGGUGCUGGUGCUCCUGGUGCCGGCGCUGGCCCAG ....((.(((((....)))))))((((((((...((((------((....))))))))))...((((((((..(.....)..))))))))))))... ( -44.40) >DroEre_CAF1 2191 94 + 1 AAUCACUCGUCGGUGGCGGCGGUGGCCAACUUUGGAUCCGGGUCCGGAUACGGAUCGGGAUCUGGGUCU---GCUCCUGGUGCUGGUGCUGGCCCAG ..((((.(((((....)))))))))........((((((.((((((....)))))).))))))(((((.---((.((.......)).)).))))).. ( -46.10) >DroAna_CAF1 2878 82 + 1 AGUGGCAUGUCUGGGUCAGGA---------UUUGGUUCCAUUGCCGGUU---CAUCUGGAGCUGGUUUU---GGUUCUGGUCCUAGUCCUGGCUCAA ...........((((((((((---------((.((..(((..(((((((---(.....))))))))..)---)).......)).)))))))))))). ( -35.90) >consensus AAUCACUCGUCGGUGGCGGCGGUGGCC__CUUUGGAUCC____CCGGAUACGGAUCGGGAUCUGGUUCU___GCUCCUGGUGCUGGUGCUGGCCCAG ........(((((((.((((..........((((((......)))))).....(((((((.(..........).))))))))))).))))))).... (-20.09 = -20.43 + 0.34)



| Location | 10,233,870 – 10,233,970 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -25.65 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10233870 100 - 22224390 UCGGCCGAUAUCAUCGUCUCGCUGGGCGGCGAUAUCGAGUAUUGCUGGAGCAGACGGCGCUGCUGUUCGGUGAGAUCGUCGAGCGGAUCAUGG-CUGGAUC-- (((((((((.((..(((((((((((((((((((((...))))))))..(((((......)))))))))))))))).))..))....)))..))-))))...-- ( -41.80) >DroVir_CAF1 2688 96 - 1 UCAGCCGAUAUCAUUGUCUCGCUGGGCGGCGAUAUCGAGUACUGUUGCAGCAUGCGGCGCUGCUGUUCGGUGAGCUCGUCCAGCUGAUCGCUUGCC------- .((((.((((....))))..))))(((((((((..(((((((((..((((((.((...)))))))).))))..)))))........))))).))))------- ( -36.20) >DroPse_CAF1 22475 97 - 1 UCGGCCGAUAUCAUCGUUUCGCUGGGCGGCGAUAUCGAAUACUGUUGCAGCAGGCGUCGCUGCUGUUCGGUCAGAUCCUCCAGCGGAUCGUGA-UUAC----- ..((((((.....(((..(((((....)))))...)))........(((((((......))))))))))))).(((((......)))))....-....----- ( -36.70) >DroWil_CAF1 192 102 - 1 UCCGCUGAGAUCAUUGUUUCGCUGGGCGGCGAUAUUGAGUAUUGUUGCAAUAGACGACGCUGCUGCUCGGUAAGAUCGUCCAAUGGAUCUGAG-CUAGAACCA ...(((.((((((((((((..(((((((((.......((..(((((......)))))..)))))))))))..))).....)))).))))).))-)........ ( -34.01) >DroMoj_CAF1 2745 96 - 1 UCGGCCGAUAUCAUUGUCUCGCUGGGCGGCGAUAUCGAGUAUUGCUGCAGCAUGCGGCGCUGCUGUUCGGUGAGCUCGUCCAGCGGAUCGUUUGCU------- .(((((((((....))))..((((((((((..(((((((((.((((((.....)))))).)))...)))))).)).)))))))))).)))......------- ( -40.90) >DroPer_CAF1 2126 97 - 1 UCGGCCGAUAUCAUCGUUUCGCUGGGCGGCGAUAUCGAAUACUGUUGCAGCAGGCGUCGCUGCUGUUCGGUCAGAUCCUCCAGCGGAUCGUGA-UUGG----- ..((((((.....(((..(((((....)))))...)))........(((((((......))))))))))))).(((((......)))))....-....----- ( -36.70) >consensus UCGGCCGAUAUCAUCGUCUCGCUGGGCGGCGAUAUCGAGUACUGUUGCAGCAGGCGGCGCUGCUGUUCGGUGAGAUCGUCCAGCGGAUCGUGA_CU_G_____ .....(((((((.(((((......))))).)))))))..(((((..(((((((......))))))).))))).((((........)))).............. (-25.65 = -25.77 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:19 2006