| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,233,666 – 10,233,760 |
| Length | 94 |
| Max. P | 0.728345 |

| Location | 10,233,666 – 10,233,760 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.84 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -18.55 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10233666 94 + 22224390 UUAAAAAACAAAAAGAGGCCGAAGAGCACGCCCAGAAGCUGCUAAUCGCUCAACAAUUGGCCGCCGCCGGCGG---AG-CAU-----CCGGCGCAU---CUGCCAC------------ ..............(((..(((..((((.((......))))))..)))))).......(((....)))(((((---((-(..-----.....)).)---)))))..------------ ( -27.20) >DroVir_CAF1 2525 115 + 1 UCAAAAAACAAAAAGAGGCCGAGGAGCAUGCACAGAAGCUGAUCAUUGCUCAACAAUUGGCCGCUUCUGGCGGCAGCGGCAGCGACGGCGGCGUCU---CUGCCUCGAGCUCAGCUGC ........((..(((.((((((.(((((((..(((...)))..)).))))).....)))))).))).))(((((((((((((.((((....)))).---)))))....)))..))))) ( -42.00) >DroGri_CAF1 2943 97 + 1 UCAAAAAACAAAAAGAGGCCGAGGAGCAUGCCCAGAAGCUGCUCAUCUCUCAACAAUUGGCUGCCGCCGGUGU---CGG---------------UG---CCGCCUCCACUUCAGCUGC ..............(((((.((((((((.((......)))))))..))).........(((.(((((....).---)))---------------))---)))))))............ ( -28.20) >DroYak_CAF1 2012 94 + 1 UUAAAAAACAAAAAGAGGCCGAAGAGCACGCCCAGAAGCUGCUAAUCGCUCAACAAUUGGCCGCCGCCGGCGG---AG-CAA-----CCGGCUUAU---CCGCCUC------------ ..............(((((.((.((((..(((..(..((.((((((.(.....).)))))).))..).)))((---..-...-----)).)))).)---).)))))------------ ( -28.10) >DroMoj_CAF1 2555 112 + 1 UCAAAAAACAAAAAGAGGCCGAGGAGCAUGCACAGAAGCUGCUCAUCGCUCAGCAAUUGGCCGCUGCUGGCGG---CGGCAGUGGCAGCGGCGUCU---CUGCCUCCAGUUCGGCCGC ................((((((((((...(((.(((.((((((....((...)).....((((((((((....---)))))))))))))))).)))---.)))))))...)))))).. ( -51.40) >DroAna_CAF1 2741 102 + 1 UCAAAAAGCAAAAAGAGGCGGAAGAGCACGCCCAGAAGCUGCUCAUUGCCCAGCAGUUGGCGGCUGCCGGCGG---UG-CAG-----CUGCCGCAGUGGCUGCCGCAUCCG------- .......((.....(.((((........)))))...(((((((........)))))))((((((..(((((((---..-...-----))))))..)..)))))))).....------- ( -43.50) >consensus UCAAAAAACAAAAAGAGGCCGAAGAGCACGCCCAGAAGCUGCUCAUCGCUCAACAAUUGGCCGCCGCCGGCGG___CG_CAG_____CCGGCGUAU___CUGCCUC_A__U_______ ..............(((((.((.(((((.((......))))))).)).............(((((...)))))............................)))))............ (-18.55 = -19.00 + 0.45)

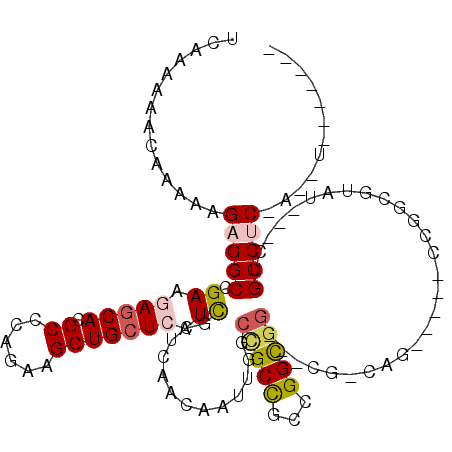

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:17 2006