
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,212,030 – 10,212,310 |
| Length | 280 |
| Max. P | 0.993953 |

| Location | 10,212,030 – 10,212,150 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -49.66 |
| Consensus MFE | -43.90 |
| Energy contribution | -43.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10212030 120 - 22224390 CCCGAAGUGUGGAUGUGGCGGUCUUCCUGCGCCAACGGAUCGCCGCCUGGUUCGAUGAGAAUCGGGAUGCGACCAGUGGCGACAUGGAGGACUAUCAGAUGCGCGGUGGACCGUAAGUAC .(((...((((.((.((..(((((((((((((((..((.((((..((((((((.....))))))))..))))))..))))).)).))))))))..)).)).)))).)))........... ( -57.30) >DroSec_CAF1 8375 120 - 1 CCCGGAGUGUGGAUGUGGCGGUCUUCCUGCGCCAGCGGAUCGCCGCCUGGUUCGAUGAGAAUCGGGAUGCGAGCAGUGGCGACAUGGAGAACUAUCAGAUGCGCGGUGGACCGUAAGUAC ..((((.((((.((.((..(((..((((((((((.(...((((..((((((((.....))))))))..))))...)))))).)).)))..)))..)).)).)))).)...)))....... ( -48.80) >DroSim_CAF1 8844 120 - 1 CCCGGAGUGUGGAUGUGGCGGUCUUCCUGCGCCAGCGGAUCGCCGCCUGGUUCGAUGAGAAUCGGGAUGCGAGCAGUGGCGACAUGGAGAACUAUCAGAUGCGCGGUGGACCGUAAGUAC ..((((.((((.((.((..(((..((((((((((.(...((((..((((((((.....))))))))..))))...)))))).)).)))..)))..)).)).)))).)...)))....... ( -48.80) >DroEre_CAF1 9523 120 - 1 CCCGGAGCGUGGAUGUGGCGGUCUUUCUGCGCCAGAGGAUCGCCGCCUGGUUCGAUGAGAAUCGGUAUGCGAGCAGUGGCGACAUGGAGAACUACCAGAUGCGCGGGGGACCGUAAGUGC ......(((((.((.(((..(((((..(((((((..(..((((...(((((((.....)))))))...)))).)..))))).))..))).))..))).)).)))((....))......)) ( -47.20) >DroYak_CAF1 9521 120 - 1 CCCGGAGCGUGGAUGUGGCGGUCUUUCUGCGCCAGAGAAUCGCCGCCUGGUUCGAUGAGAAUCGGGAUGCGACCAGUGGCGACAUUGAGAGCUACCAGAUGCGUGGAGGACCGUAAGUUC ..........((((...((((((((.(..(((.......((((..((((((((.....))))))))..)))).(((((....))))).............)))..)))))))))..)))) ( -46.20) >consensus CCCGGAGUGUGGAUGUGGCGGUCUUCCUGCGCCAGCGGAUCGCCGCCUGGUUCGAUGAGAAUCGGGAUGCGAGCAGUGGCGACAUGGAGAACUAUCAGAUGCGCGGUGGACCGUAAGUAC ........(((.((.(((..((((((.(((((((.....((((..((((((((.....))))))))..))))....))))).)).)))).))..))).)).)))((....))........ (-43.90 = -43.62 + -0.28)



| Location | 10,212,110 – 10,212,230 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -44.36 |
| Energy contribution | -44.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10212110 120 + 22224390 AUCCGUUGGCGCAGGAAGACCGCCACAUCCACACUUCGGGUGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGC ..(((((((((((.((.(((.(((((((((.......)))))..(((....)))......)))).)((...((((((((....)))))))).))..)).)).))))))).))))...... ( -49.40) >DroSec_CAF1 8455 120 + 1 AUCCGCUGGCGCAGGAAGACCGCCACAUCCACACUCCGGGUGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGC ..(((((((((((.((.(((.(((((((((.......)))))..(((....)))......)))).)((...((((((((....)))))))).))..)).)).))))))).))))...... ( -51.80) >DroSim_CAF1 8924 120 + 1 AUCCGCUGGCGCAGGAAGACCGCCACAUCCACACUCCGGGUGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGC ..(((((((((((.((.(((.(((((((((.......)))))..(((....)))......)))).)((...((((((((....)))))))).))..)).)).))))))).))))...... ( -51.80) >DroEre_CAF1 9603 120 + 1 AUCCUCUGGCGCAGAAAGACCGCCACAUCCACGCUCCGGGUGAAGAGAAUGCUCAGAUUCUGGCCCGCAUAGCGAUACGUGUUCGUGUUCCGGCAUUCAUCGUGCGCCAUGCGGCACUGC ..........((((.......(((((((((.......)))))..(((....)))......))))((((((.(((.((((((...((((....)))).)).))))))).))))))..)))) ( -38.70) >DroYak_CAF1 9601 120 + 1 AUUCUCUGGCGCAGAAAGACCGCCACAUCCACGCUCCGGGUGAACAGGAUGCUCAGAUUCUGGCCCGCAUAGCGAUAUGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGC ......((((((.....(((((.(((((...((((.(((((...((((((......)))))))))))...))))..)))))..)).)))(((((....))))))))))).(((....))) ( -41.30) >consensus AUCCGCUGGCGCAGGAAGACCGCCACAUCCACACUCCGGGUGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGC ..(((((((((((.((.((..(((.(((((.......)))))...(((((......))))))))..((...((((((((....)))))))).))..)).)).))))))).))))...... (-44.36 = -44.72 + 0.36)

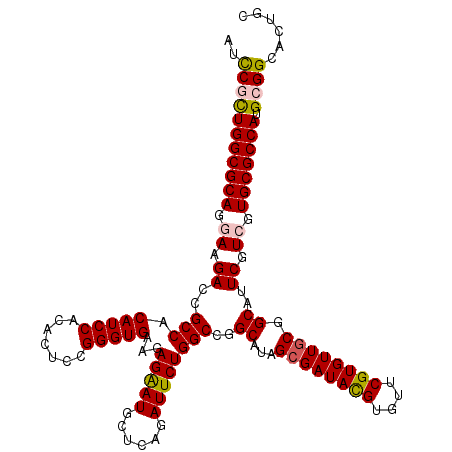

| Location | 10,212,150 – 10,212,270 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -50.36 |
| Consensus MFE | -42.88 |
| Energy contribution | -42.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10212150 120 + 22224390 UGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGCAGCGCAUUGUAGGCGGCCAGUUGGGCCAGUGUCGUGUCCC ....(((....))).(((.(((((((((...((((((((....)))))))).((((.....)))))))..((.((.((((((....)))))))).))......)))))).)))....... ( -49.40) >DroSec_CAF1 8495 120 + 1 UGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGCAGCGCAUUGUAUGCGGCCAGUUGGGCCAGUGUCGUGUCCC .((.((.(..((((((...(((((((.((((((((((((....))))))))(((....)))((((((..((((....))))))))))..)))))))))))))))))...).))...)).. ( -49.60) >DroSim_CAF1 8964 120 + 1 UGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGCAGCGCAUUGUAUGCGGCCAGUUGGGCCAGUGUCGUGUCCC .((.((.(..((((((...(((((((.((((((((((((....))))))))(((....)))((((((..((((....))))))))))..)))))))))))))))))...).))...)).. ( -49.60) >DroEre_CAF1 9643 120 + 1 UGAAGAGAAUGCUCAGAUUCUGGCCCGCAUAGCGAUACGUGUUCGUGUUCCGGCAUUCAUCGUGCGCCAUGCGGCACUGCAGCGCAUUGUAGGCGGCCAAUUGGGCCAGUGUGGCGUCCC ........((((.((.(..(((((((((((.(((.((((((...((((....)))).)).))))))).))))(((.((((((....))))))...)))....)))))))).))))))... ( -46.80) >DroYak_CAF1 9641 120 + 1 UGAACAGGAUGCUCAGAUUCUGGCCCGCAUAGCGAUAUGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGCAGCGCAUUGUAGGCGGCCAGUUGGGCCAGUGUCGCAUCCC ......((((((...(((.(((((((((...((((((((....))))))))(((...((((((((((..((((....))))))))))....))))))).).)))))))).))))))))). ( -56.40) >consensus UGAAGAGAAUGCUCAGAUUCUGGCCGGCAUAGCGAUACGUGUUCGUGUUGCGGCAUUCGUCGUGCGCCAUGCGGCACUGCAGCGCAUUGUAGGCGGCCAGUUGGGCCAGUGUCGUGUCCC .....(((((......)))))(((..((((.((((((((....))))))))(((....))))))))))((((((((((.....((.......))((((.....))))))))))))))... (-42.88 = -42.76 + -0.12)


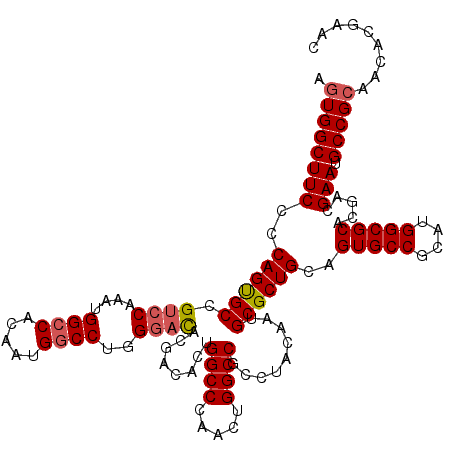
| Location | 10,212,190 – 10,212,310 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -47.58 |
| Consensus MFE | -43.80 |
| Energy contribution | -44.28 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10212190 120 - 22224390 AGUGGCUUCCCCAGUGCCGUCCAAAUGGCCACAAUGUCCUGGGACACGACACUGGCCCAACUGGCCGCCUACAAUGCGCUGCAGUGCCGCAUGGCGCACGACGAAUGCCGCAACACGAAC .((((((((..((((((.((((....((.(.....).))..))))........((((.....)))).........))))))..(((((....))))).....))).)))))......... ( -41.70) >DroSec_CAF1 8535 120 - 1 AGUGGCUUCCCCAGUGCCGGCCAAAUGGCCACAAUGGCCUGGGACACGACACUGGCCCAACUGGCCGCAUACAAUGCGCUGCAGUGCCGCAUGGCGCACGACGAAUGCCGCAACACGAAC .((((((((..((((((((((((...((((.....))))((((.((......)).))))..))))))........))))))..(((((....))))).....))).)))))......... ( -49.40) >DroSim_CAF1 9004 120 - 1 AGUGGCUUCCCCAGUGCCGUCCAAAUGGCCACAAUGGCCUGGGACACGACACUGGCCCAACUGGCCGCAUACAAUGCGCUGCAGUGCCGCAUGGCGCACGACGAAUGCCGCAACACGAAC .((((((((..((((((.((((....((((.....))))..))))........((((.....)))).........))))))..(((((....))))).....))).)))))......... ( -48.80) >DroEre_CAF1 9683 120 - 1 AGUGGCUUCCCCAGCGCCGUCCAGAUGGCCACAAUGGCCUGGGACGCCACACUGGCCCAAUUGGCCGCCUACAAUGCGCUGCAGUGCCGCAUGGCGCACGAUGAAUGCCGGAACACGAAC .(((((...(((((.(((((((....)).....))))))))))..))))).(((((...((((..((((....((((((......).)))))))))..))))....)))))......... ( -48.70) >DroYak_CAF1 9681 120 - 1 AGUGGCUUCCCCAGUGCCGUGCAAAUGGCCACAAUGGCCUGGGAUGCGACACUGGCCCAACUGGCCGCCUACAAUGCGCUGCAGUGCCGCAUGGCGCACGACGAAUGCCGCAACACGAAC .((((((((....(((((((((....((((.....))))..((.((((.....((((.....))))(((......).))))))...))))))))))).....))).)))))......... ( -49.30) >consensus AGUGGCUUCCCCAGUGCCGUCCAAAUGGCCACAAUGGCCUGGGACACGACACUGGCCCAACUGGCCGCCUACAAUGCGCUGCAGUGCCGCAUGGCGCACGACGAAUGCCGCAACACGAAC .((((((((..((((((.((((....((((.....))))..))))........((((.....)))).........))))))..(((((....))))).....))).)))))......... (-43.80 = -44.28 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:08 2006