
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,210,281 – 10,210,392 |
| Length | 111 |
| Max. P | 0.987720 |

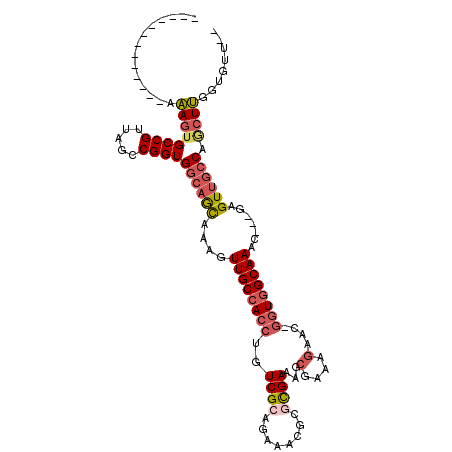
| Location | 10,210,281 – 10,210,378 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -25.58 |
| Energy contribution | -26.62 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10210281 97 + 22224390 -------------AAAGUGCCGUUAGCCGGUGGCAACAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAAC---GAGUUGCCAGCUUGUUGUUUC -------------.((((((((.....))))((((((....((((((((..((((......))((....))....))..-))))))))..---..)))))).))))........ ( -39.30) >DroSec_CAF1 6616 93 + 1 -------------AAAGUGCCGUUAGCCGGUGGCAGCAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAAC---GAGUUGCCAGCUUGUUG---- -------------.((((((((.....))))((((((....((((((((..((((......))((....))....))..-))))))))..---..)))))).))))....---- ( -39.30) >DroSim_CAF1 7069 97 + 1 -------------AAAGUGCCGUUAGCCGGUGGCAGCAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAAC---GAGUUGCCAGCUUGUUGUUUC -------------.((((((((.....))))((((((....((((((((..((((......))((....))....))..-))))))))..---..)))))).))))........ ( -39.30) >DroEre_CAF1 7781 93 + 1 -------------AAAGUGCCGUUAGCCGGUGGCAGCAAAGUUGCCACCUGUCGA--ACUCGCGCGAUAGCGAAAGAAC-GUUGGCAAAG---GAGUUGCCAACUUGGUGAU-- -------------.....((((((.((.(((((((((...))))))))).))...--.((..(((....)))..)))))-((((((((..---...))))))))..)))...-- ( -37.10) >DroYak_CAF1 7727 98 + 1 -------------AAAGUGCCGUUAGCCGGUGGCAGCAAUGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAACGACGAGUUGCCAGCUUGGUAAU-- -------------....(((((..(((.((..((.......((((((((..((((......))((....))....))..-)))))))).......))..)).))))))))..-- ( -38.94) >DroPer_CAF1 9460 101 + 1 GCCAGCAGCUGAGAGAGAGCCGGUG--CGGUG--UGUAAGGUUGCCACCUGUCGGCGAAGCAGGUGAA------AAAACAAGUAGCAAAA---GUGUUGCCAGGUCUGCCUUUG (((..((.(((.......((((..(--.((((--.(((....))))))))..))))....))).))..------.......((((((...---.))))))..)))......... ( -28.70) >consensus _____________AAAGUGCCGUUAGCCGGUGGCAGCAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC_GGUGGCAAAC___GAGUUGCCAGCUUGGUGUU__ ..............((((((((.....))))((((((....((((((((..((((........))))...(....)....)))))))).......)))))).))))........ (-25.58 = -26.62 + 1.03)


| Location | 10,210,281 – 10,210,378 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.68 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10210281 97 - 22224390 GAAACAACAAGCUGGCAACUC---GUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGUUGCCACCGGCUAACGGCACUUU------------- ..........((.(....).(---(((.(((..(-((.....(((......))).....)))...(((((((((...)))))))))))).)))))).....------------- ( -34.30) >DroSec_CAF1 6616 93 - 1 ----CAACAAGCUGGCAACUC---GUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGCUGCCACCGGCUAACGGCACUUU------------- ----......((.(....).(---(((.(((..(-((.....(((......))).....)))...(((((((.......)))))))))).)))))).....------------- ( -31.50) >DroSim_CAF1 7069 97 - 1 GAAACAACAAGCUGGCAACUC---GUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGCUGCCACCGGCUAACGGCACUUU------------- ..........((.(....).(---(((.(((..(-((.....(((......))).....)))...(((((((.......)))))))))).)))))).....------------- ( -31.50) >DroEre_CAF1 7781 93 - 1 --AUCACCAAGUUGGCAACUC---CUUUGCCAAC-GUUCUUUCGCUAUCGCGCGAGU--UCGACAGGUGGCAACUUUGCUGCCACCGGCUAACGGCACUUU------------- --........((((((((...---..))))))))-(((..(((((......))))).--..))).(((((((.......)))))))(.(....).).....------------- ( -33.50) >DroYak_CAF1 7727 98 - 1 --AUUACCAAGCUGGCAACUCGUCGUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACAUUGCUGCCACCGGCUAACGGCACUUU------------- --...........(....)..((((((.(((..(-((.....(((......))).....)))...(((((((.......)))))))))).)))))).....------------- ( -34.20) >DroPer_CAF1 9460 101 - 1 CAAAGGCAGACCUGGCAACAC---UUUUGCUACUUGUUUU------UUCACCUGCUUCGCCGACAGGUGGCAACCUUACA--CACCG--CACCGGCUCUCUCUCAGCUGCUGGC ....((((((..((....)).---.)))))).........------.((((((((......).)))))))..........--.....--..(((((............))))). ( -24.70) >consensus __AACAACAAGCUGGCAACUC___GUUUGCCACC_GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGCUGCCACCGGCUAACGGCACUUU_____________ ..........((((((((........)))))............(((.(((((......)))))..(((((((.......))))))))))....))).................. (-21.60 = -22.68 + 1.08)


| Location | 10,210,302 – 10,210,392 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.32 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -13.91 |
| Energy contribution | -16.08 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10210302 90 + 22224390 AACAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAAC---GAGUUGCCAGCUUGUUGUUUCGUCA------------------UUGUUUCUGU ...................((((((((..((....))((((.(((-(.((((((..---...)))))).)..))).))))....------------------..)))))))) ( -29.40) >DroPse_CAF1 9371 103 + 1 UACAAGGUUGCCACCUGUCGGCGAAGCAGGUGGA------AAAACAAGUACCAAAA---GUGUUGCCAGGUCUGCCUUUGGUCGACCCUUUUUUGUGGCUUUUUGUUUUUGU ..........((((((((.......))))))))(------((((((((..((((((---(.(((((((((....))..))).)))).))))....)))...))))))))).. ( -31.60) >DroSec_CAF1 6637 79 + 1 AGCAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAAC---GAGUUGCCAGCUUGUUG-----------------------------UUUCUGU .(((....(((((((..((((......))((....))....))..-)))))))(((---((((.....))))))).-----------------------------....))) ( -27.40) >DroSim_CAF1 7090 90 + 1 AGCAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAAC---GAGUUGCCAGCUUGUUGUUUCGUCA------------------UUGUUUCUGU .(((....)))........((((((((..((....))((((.(((-(.((((((..---...)))))).)..))).))))....------------------..)))))))) ( -30.60) >DroEre_CAF1 7802 83 + 1 AGCAAAGUUGCCACCUGUCGA--ACUCGCGCGAUAGCGAAAGAAC-GUUGGCAAAG---GAGUUGCCAACUUGGUGAU-----G------------------UUGUUUCUGU (((((.((..(((.((((((.--.......))))))(....)...-((((((((..---...)))))))).)))..))-----.------------------)))))..... ( -27.60) >DroYak_CAF1 7748 88 + 1 AGCAAUGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC-GGUGGCAAACGACGAGUUGCCAGCUUGGUAAU-----G------------------UUGUUUCUGU .(((.....((((((..((((......))((....))....))..-))))))(((((((..((((((.....))))))-----)------------------)))))).))) ( -31.40) >consensus AGCAAAGUUGCCACCUGUCGCAGAAACGCGCGAAAGCGAAAGAAC_GGUGGCAAAC___GAGUUGCCAGCUUGGUGUU_____G__________________UUGUUUCUGU ...................((((((((((((....)))........(.((((((........)))))).).................................))))))))) (-13.91 = -16.08 + 2.17)



| Location | 10,210,302 – 10,210,392 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.32 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10210302 90 - 22224390 ACAGAAACAA------------------UGACGAAACAACAAGCUGGCAACUC---GUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGUU .(((((((..------------------....((((.(((....((((((...---..))))))..-))).))))((......))))))))).((((((.(....))))))) ( -24.70) >DroPse_CAF1 9371 103 - 1 ACAAAAACAAAAAGCCACAAAAAAGGGUCGACCAAAGGCAGACCUGGCAACAC---UUUUGGUACUUGUUUU------UCCACCUGCUUCGCCGACAGGUGGCAACCUUGUA ..((((((((...((((....((((((((..((...))..))))((....)))---)))))))..)))))))------)((((((((......).))))))).......... ( -29.20) >DroSec_CAF1 6637 79 - 1 ACAGAAA-----------------------------CAACAAGCUGGCAACUC---GUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGCU .(((...-----------------------------.......)))((((...---(((.((((((-.......(((....)))(((....)))...))))))))).)))). ( -21.50) >DroSim_CAF1 7090 90 - 1 ACAGAAACAA------------------UGACGAAACAACAAGCUGGCAACUC---GUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGCU .(((((((..------------------....((((.(((....((((((...---..))))))..-))).))))((......)))))))))........((((....)))) ( -22.60) >DroEre_CAF1 7802 83 - 1 ACAGAAACAA------------------C-----AUCACCAAGUUGGCAACUC---CUUUGCCAAC-GUUCUUUCGCUAUCGCGCGAGU--UCGACAGGUGGCAACUUUGCU ..........------------------.-----..((((..((((((((...---..))))))))-(((..(((((......))))).--..))).))))(((....))). ( -26.40) >DroYak_CAF1 7748 88 - 1 ACAGAAACAA------------------C-----AUUACCAAGCUGGCAACUCGUCGUUUGCCACC-GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACAUUGCU .(((((((..------------------.-----........(.((((((........)))))).)-.......(((....))).))))))).....((((....))))... ( -22.00) >consensus ACAGAAACAA__________________C_____AACAACAAGCUGGCAACUC___GUUUGCCACC_GUUCUUUCGCUUUCGCGCGUUUCUGCGACAGGUGGCAACUUUGCU .........................................(((.(....).......((((((((.(((....(((......))).......))).))))))))....))) (-13.69 = -14.33 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:03 2006