
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,208,521 – 10,208,622 |
| Length | 101 |
| Max. P | 0.631919 |

| Location | 10,208,521 – 10,208,622 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.46 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10208521 101 + 22224390 CAAAGUUCAAUGUAA-AAGCAUGGUAUUUUGGUUGCUUGAAUAAUACAUUUUUCUCUCUGUGUACCCAGCGCCGUGCAUUUGUAUCCGCUGUUUGCGCUGGA ((((((...((((..-..))))...)))))).......................(((..(((((..(((((..((((....)))).)))))..))))).))) ( -22.80) >DroSim_CAF1 5297 102 + 1 CAACGUUAAAUAUUACCAGCAUGGUGUUUUGUGUGUUUGGAUAUUACAUUUUUCUCUCUGUGUACCCAGCGCCGUGCAUUUGUAUCCGCUGUUUGCACUGGA ...............(((((((((((((..(((..(..(((.............)))..)..)))..))))))))))..........((.....))..))). ( -24.72) >DroYak_CAF1 5808 87 + 1 UAAAAUUUAAUGCAA---------------AAUCGCUUUGGCUAUAACAUUUUCUUUCUGUGCACCCAGCGCCGUGCACUUGUAUCCGCUGUUUGCGCUGGA ...............---------------....((....)).............(((.(((((..(((((..((((....)))).)))))..))))).))) ( -19.80) >consensus CAAAGUUAAAUGUAA__AGCAUGGU_UUUUGAUUGCUUGGAUAAUACAUUUUUCUCUCUGUGUACCCAGCGCCGUGCAUUUGUAUCCGCUGUUUGCGCUGGA ........................................................((((((((..(((((..((((....)))).)))))..))))).))) (-17.57 = -17.13 + -0.44)


| Location | 10,208,521 – 10,208,622 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 72.46 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
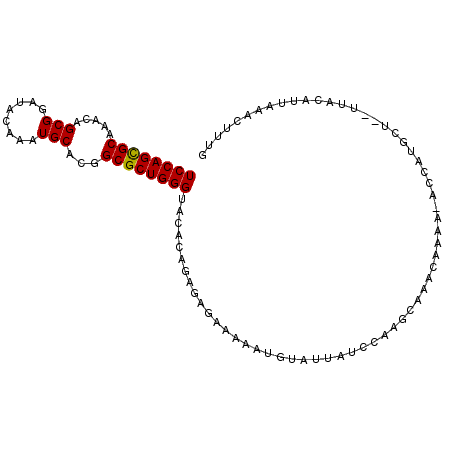
>X_DroMel_CAF1 10208521 101 - 22224390 UCCAGCGCAAACAGCGGAUACAAAUGCACGGCGCUGGGUACACAGAGAGAAAAAUGUAUUAUUCAAGCAACCAAAAUACCAUGCUU-UUACAUUGAACUUUG ((((((((.....(((........)))...))))))))....(((((.....((((((......(((((............)))))-.))))))...))))) ( -24.20) >DroSim_CAF1 5297 102 - 1 UCCAGUGCAAACAGCGGAUACAAAUGCACGGCGCUGGGUACACAGAGAGAAAAAUGUAAUAUCCAAACACACAAAACACCAUGCUGGUAAUAUUUAACGUUG ((((((((.....(((........)))...))))))))..............(((((((((((((...................)))..)))))..))))). ( -16.71) >DroYak_CAF1 5808 87 - 1 UCCAGCGCAAACAGCGGAUACAAGUGCACGGCGCUGGGUGCACAGAAAGAAAAUGUUAUAGCCAAAGCGAUU---------------UUGCAUUAAAUUUUA ((((((((.....((........)).....))))))))......((((...(((((....((....))....---------------..)))))...)))). ( -20.70) >consensus UCCAGCGCAAACAGCGGAUACAAAUGCACGGCGCUGGGUACACAGAGAGAAAAAUGUAUUAUCCAAGCAAACAAAA_ACCAUGCU__UUACAUUAAACUUUG ((((((((.....(((........)))...))))))))................................................................ (-15.59 = -15.37 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:59 2006