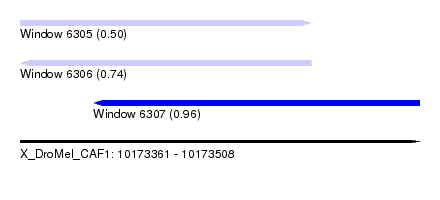
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,173,361 – 10,173,508 |
| Length | 147 |
| Max. P | 0.962922 |

| Location | 10,173,361 – 10,173,468 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -16.87 |
| Energy contribution | -16.75 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10173361 107 + 22224390 ------UAUA-------UUCUGUGGUUUGGCCAACUGUGUGUUCUAGUUUUCCACUAAUGCCCUUUAUAUUAAAUAAUAAGAACAUUAUCAAUUUAAUGAGGGCAUGCAUUAUCAGCUUA ------....-------.....(((.....))).(((.((((..((((.....))))((((((((...(((((((.((((.....))))..)))))))))))))))))))...))).... ( -21.60) >DroSec_CAF1 1517 107 + 1 ------UAUA-------CUCUGUGGUUUGGCCAACUCUGUGUUCCAGUUUUCCGCUAAUGCCCUUUAUAUUAAAUAAUAAGAACAUUAUCAAUUUAAUGAGGGCAUGCAUUAUCAGCUAA ------....-------....((((.((((..(((.....)))))))....))))..((((((((...(((((((.((((.....))))..)))))))))))))))((.......))... ( -22.50) >DroSim_CAF1 5193 107 + 1 ------UAUA-------UUCUGUGGUUUGGCCAACUCUGUGUUCUAGUUUUCCGCUAAUGCCCUUUAUAUUAAAUAAUAAGAACAUUAUCAAUUUAAUGAGGGCAUGCAUUAUCAGCUAA ------.(((-------(....(((.....))).....))))..(((((........((((((((...(((((((.((((.....))))..)))))))))))))))........))))). ( -20.49) >DroEre_CAF1 7687 116 + 1 CUUAGUUAUAUUUAUGCUUCUGCGG-UUGGACUUCUAUGUGCUUUAGUUUUCUGCCAAUGCCCUUUAUAUUAAAUAAUA---ACACUAUCUUUUUAAUGAGGGCAUGCAUUAUCAGCUUA ...............(((..(((((-(.(((...(((.......)))..))).))).((((((((...((((((..(((---....)))...))))))))))))))))).....)))... ( -22.30) >DroYak_CAF1 4841 114 + 1 CUUCGUUAUAUUUUU------GCGGUUUGGACUUCUAUGUGUUCUAGUUUUCUGCCAAUGCCCUUUGUAUUAGAUAAUAAGAACACUAUCUAUUUAAUGAGGGCAUGCAUUAUCAGCAUA ....((((((....(------(((((..(((((............)))))...))).((((((((.....((((((..........))))))......))))))))))).))).)))... ( -26.20) >consensus ______UAUA_______UUCUGUGGUUUGGCCAACUAUGUGUUCUAGUUUUCCGCUAAUGCCCUUUAUAUUAAAUAAUAAGAACAUUAUCAAUUUAAUGAGGGCAUGCAUUAUCAGCUUA .....................((((.((((..(((.....)))))))....))))..((((((((...(((((((................)))))))))))))))((.......))... (-16.87 = -16.75 + -0.12)


| Location | 10,173,361 – 10,173,468 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -15.88 |
| Energy contribution | -15.08 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10173361 107 - 22224390 UAAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGUGGAAAACUAGAACACACAGUUGGCCAAACCACAGAA-------UAUA------ ....(((.......(((((((..(((.((((((((((....))))).))))).))))))))))...(((..((((.(......)))))..))).....)))..-------....------ ( -21.20) >DroSec_CAF1 1517 107 - 1 UUAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGCGGAAAACUGGAACACAGAGUUGGCCAAACCACAGAG-------UAUA------ ...((..((..((((((((((..(((.((((((((((....))))).))))).))))))))))..))).....(((.....))).))..))............-------....------ ( -23.30) >DroSim_CAF1 5193 107 - 1 UUAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGCGGAAAACUAGAACACAGAGUUGGCCAAACCACAGAA-------UAUA------ ....(((.....(((((((((..(((.((((((((((....))))).))))).))))))))))..))((..((((.........))))..))......)))..-------....------ ( -22.70) >DroEre_CAF1 7687 116 - 1 UAAGCUGAUAAUGCAUGCCCUCAUUAAAAAGAUAGUGU---UAUUAUUUAAUAUAAAGGGCAUUGGCAGAAAACUAAAGCACAUAGAAGUCCAA-CCGCAGAAGCAUAAAUAUAACUAAG ...(((.....(((((((((((((((......))))).---..((((.....))))))))))).((..((...(((.......)))...))...-)))))..)))............... ( -19.30) >DroYak_CAF1 4841 114 - 1 UAUGCUGAUAAUGCAUGCCCUCAUUAAAUAGAUAGUGUUCUUAUUAUCUAAUACAAAGGGCAUUGGCAGAAAACUAGAACACAUAGAAGUCCAAACCGC------AAAAAUAUAACGAAG ..(((......((((((((((.......(((((((((....)))))))))......)))))))..)))((...(((.......)))...))......))------).............. ( -23.52) >consensus UAAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGCGGAAAACUAGAACACAGAGUUGGCCAAACCACAGAA_______UAUA______ ...(((((.......((((((.........(((((((....)))))))........)))))))))))..................................................... (-15.88 = -15.08 + -0.80)


| Location | 10,173,388 – 10,173,508 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.31 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10173388 120 - 22224390 UAUCUUUAUAAUUGCACUUAACACAAUUAUAAACACUUACUAAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGUGGAAAACUAGAAC ....((((((((((.........))))))))))...(((((((((.......)).((((((..(((.((((((((((....))))).))))).))))))))))))))))........... ( -23.80) >DroSec_CAF1 1544 120 - 1 UAUCUUUAUAAUUGCACUUAACACAAUUAUAAACACUGACUUAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGCGGAAAACUGGAAC ....((((((((((.........))))))))))..(((.((..((.......))(((((((..(((.((((((((((....))))).))))).)))))))))).)))))........... ( -22.00) >DroSim_CAF1 5220 120 - 1 UAUCUUUAUAAUUGCACUUAACACAAUUAUAAACACUGACUUAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGCGGAAAACUAGAAC ....((((((((((.........))))))))))..(((.((..((.......))(((((((..(((.((((((((((....))))).))))).)))))))))).)))))........... ( -22.00) >DroEre_CAF1 7726 117 - 1 UAUCAUGCUAAUUGCACUUAACACGAAUAUAACCACUGACUAAGCUGAUAAUGCAUGCCCUCAUUAAAAAGAUAGUGU---UAUUAUUUAAUAUAAAGGGCAUUGGCAGAAAACUAAAGC .....(((.....)))...................(((.((((((.......)).(((((((((((......))))).---..((((.....)))))))))))))))))........... ( -19.00) >DroYak_CAF1 4875 120 - 1 CAUCAUUCUAAUUGCACUUAACACGAGUAUAAACACUGACUAUGCUGAUAAUGCAUGCCCUCAUUAAAUAGAUAGUGUUCUUAUUAUCUAAUACAAAGGGCAUUGGCAGAAAACUAGAAC .....(((((.(((.((((.....)))).)))...(((.((((((.......)))((((((.......(((((((((....)))))))))......)))))).)))))).....))))). ( -26.72) >consensus UAUCUUUAUAAUUGCACUUAACACAAUUAUAAACACUGACUAAGCUGAUAAUGCAUGCCCUCAUUAAAUUGAUAAUGUUCUUAUUAUUUAAUAUAAAGGGCAUUAGCGGAAAACUAGAAC .....(((((((((.........)))))))))...(((.(((.((.......))(((((((.........(((((((....)))))))........)))))))))))))........... (-18.83 = -19.31 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:51 2006