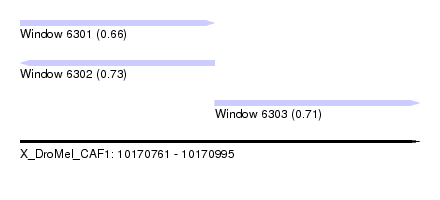
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,170,761 – 10,170,995 |
| Length | 234 |
| Max. P | 0.729370 |

| Location | 10,170,761 – 10,170,875 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -28.61 |
| Energy contribution | -29.61 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10170761 114 + 22224390 CUGAUGGGGGAGAGCGGGAAAAAGGAGGUGACGAA----AGAGAGAGGCAACAGAAA-AAGGGAUGAAUCUUUAGAUUUGGAUUUGAUUUUUCUGACUUCUCUUCGCUCUCCUU-UCCCA ....((((((((((((..........(....)...----.((((((((...((((((-((....(((((((........))))))).)))))))).))))))))))))))))..-.)))) ( -41.40) >DroSim_CAF1 2294 113 + 1 CUGAUGGGGGAGAGCGGGAAAAAGGAGGUGACGAA----AGA--GAGGCAACAGAAAAAAGGGAUGAAUCUUUAGAUUUGGAUUUGAUUUUUCUGUCUUCUCUCCGCUCUCCUU-UCCCA ....(((((((((((((.........(....)...----(((--((((..(((((((((..(((((((((....)))))..))))..)))))))))))))))))))))))))..-.)))) ( -44.70) >DroEre_CAF1 5304 113 + 1 CUGAUGG-GGAGAGUGGGAAAAAGGAGGUGACGAA----AGAGAGAGGAAACAGAAA-AAGGGAUGAAUCUUUAGAUUUGGAUUUGAUUUUUCUGCCUUCUCUUCGUUCUCCUU-UCCAC .......-.....((((((...(((((..((((((----.((((.(((...((((((-((....(((((((........))))))).)))))))))))))))))))))))))))-))))) ( -36.00) >DroYak_CAF1 2345 118 + 1 CUGAUAG-GAAGAGUGGGAAAAAGGAGGUGACGAAAGUAAGAGAGAGGAAACAGAAA-AAGGGAUGAAUCUUUAGAUUUGGAUUUGAUUUUUCUGUCUUCUCUCCGUUCUCCUUUUCCCC .......-.......(((((((.(((((..((....))..(((((((((..((((((-((....(((((((........))))))).)))))))))))))))))...)))))))))))). ( -41.70) >consensus CUGAUGG_GGAGAGCGGGAAAAAGGAGGUGACGAA____AGAGAGAGGAAACAGAAA_AAGGGAUGAAUCUUUAGAUUUGGAUUUGAUUUUUCUGUCUUCUCUCCGCUCUCCUU_UCCCA ...............((((...((((((...((.......((((((((..(((((((..(((..((((((....))))))..)))....))))))))))))))))).))))))..)))). (-28.61 = -29.61 + 1.00)


| Location | 10,170,761 – 10,170,875 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10170761 114 - 22224390 UGGGA-AAGGAGAGCGAAGAGAAGUCAGAAAAAUCAAAUCCAAAUCUAAAGAUUCAUCCCUU-UUUCUGUUGCCUCUCUCU----UUCGUCACCUCCUUUUUCCCGCUCUCCCCCAUCAG ((((.-..((((((((.(((((.((((((((((.........((((....))))......))-))))))..)).)))))..----...(......)........)))))))))))).... ( -29.56) >DroSim_CAF1 2294 113 - 1 UGGGA-AAGGAGAGCGGAGAGAAGACAGAAAAAUCAAAUCCAAAUCUAAAGAUUCAUCCCUUUUUUCUGUUGCCUC--UCU----UUCGUCACCUCCUUUUUCCCGCUCUCCCCCAUCAG ((((.-..(((((((((((((..((((((((((.........((((....)))).......))))))))))..)))--)).----...(......)........)))))))))))).... ( -36.79) >DroEre_CAF1 5304 113 - 1 GUGGA-AAGGAGAACGAAGAGAAGGCAGAAAAAUCAAAUCCAAAUCUAAAGAUUCAUCCCUU-UUUCUGUUUCCUCUCUCU----UUCGUCACCUCCUUUUUCCCACUCUCC-CCAUCAG (((((-((((((.((((((((((((((((((((.........((((....))))......))-))))))...))).)))).----)))))...)))))))...)))).....-....... ( -30.66) >DroYak_CAF1 2345 118 - 1 GGGGAAAAGGAGAACGGAGAGAAGACAGAAAAAUCAAAUCCAAAUCUAAAGAUUCAUCCCUU-UUUCUGUUUCCUCUCUCUUACUUUCGUCACCUCCUUUUUCCCACUCUUC-CUAUCAG ((((((((((((...((((((.(((((((((((.........((((....))))......))-))))))))).))))))...((....))...)))))))))))).......-....... ( -37.16) >consensus GGGGA_AAGGAGAACGAAGAGAAGACAGAAAAAUCAAAUCCAAAUCUAAAGAUUCAUCCCUU_UUUCUGUUGCCUCUCUCU____UUCGUCACCUCCUUUUUCCCACUCUCC_CCAUCAG .((((..(((((.((((((((.(((((((((...........((((....)))).........))))))))).))).........)))))...)))))...))))............... (-24.75 = -25.12 + 0.38)

| Location | 10,170,875 – 10,170,995 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -37.25 |
| Consensus MFE | -31.42 |
| Energy contribution | -32.48 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10170875 120 + 22224390 CACUAGCAUCCAUCCAGCUACCUCUAUCUCGCUUCCACCUUAGCCGCUUCUUCCAGUUGUUUUUGUUGUUACUUUCCGUUCCUGGCAGGAGGUGUGUUUGUGCGAGAGAGAGGGGAAUGC .....((((...(((.....(((((.((((((...((....(((((((((((((((........((....)).........)))).)))))))).))))).)))))).)))))))))))) ( -35.63) >DroSim_CAF1 2407 120 + 1 CACUAGCAUCCAUCCUGCUAACUCCAUCUCGCUUCCACCUUAGCCGCUUCUUCCAGUUGUUUUUGUUGUUACUUUCCGUUCCUGGCAGGAGGUGUGUUUGUGCGAGCGAGAGGGGAAUGA ...(((((.......))))).((((.((((((((.(((...(((((((((((((((........((....)).........)))).)))))))).))).))).))))))))))))..... ( -42.03) >DroEre_CAF1 5417 120 + 1 CGCUGGCAUCCAUCUCGGUGUCUCCAUCUCGCUUGCACCUUAGCCGCUUCUUUCAGUUGUUUUUGUUGUUACUUUCCGUUCCUCGCAGGAGGUGUGUUUGUGCGAGCGAGAAGAGAAUAC ((((((........))))))((((..((((((((((((...(((((((((((.(((........((....)).........)).).)))))))).))).)))))))))))).)))).... ( -39.23) >DroYak_CAF1 2463 120 + 1 CACUAGCAUCCAUCUAGGUAUCUCCAUCUCACUUGCGCCUUAGCCGCUUCUUUCAGUUGUUUUUGUUGUUGCUAUCCGUUCCUCGCAGGAGGUGUGUUUGUGCGAGCGAGACGAGAAUAC ..((((.......))))(((((((..((((.(((((((...((((((((((..(((......)))....(((............)))))))))).))).))))))).)))).))).)))) ( -32.10) >consensus CACUAGCAUCCAUCCAGCUAUCUCCAUCUCGCUUCCACCUUAGCCGCUUCUUCCAGUUGUUUUUGUUGUUACUUUCCGUUCCUCGCAGGAGGUGUGUUUGUGCGAGCGAGAGGAGAAUAC .....................((((.((((((((((((...(((((((((((((((........((....)).........)))).)))))))).))).))))))))))))))))..... (-31.42 = -32.48 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:47 2006