
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,164,431 – 10,164,572 |
| Length | 141 |
| Max. P | 0.998279 |

| Location | 10,164,431 – 10,164,536 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
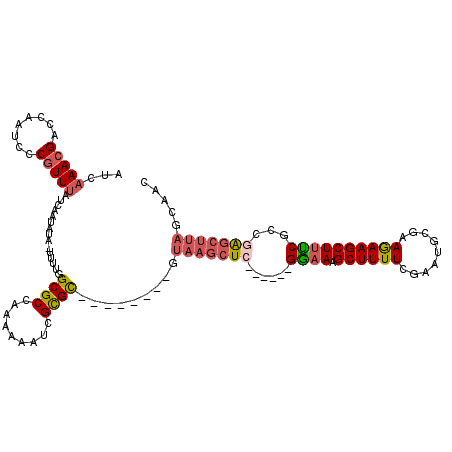
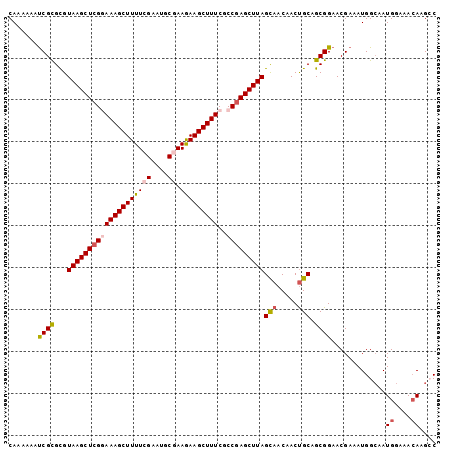
>X_DroMel_CAF1 10164431 105 + 22224390 AUCAAACGACCAAUCGCGUUUAUCAAUAUA-UUUUGGCGCCAAAAAAUCGCGC--------GUAAGCUC------GGAAAGCUUUUCGAAUGCGAAGAAGCUUUCGCCAAGCUUAGCAAC ...(((((........))))).........-.((((((..........((.((--------....)).)------)((((((((((((....)).))))))))))))))))......... ( -29.30) >DroPse_CAF1 32973 110 + 1 AACUAACGCCGCAAAACAUUCAACAAUCUUUUUUUGGCGCCAAAAAAUCGCGUAAGCGUAGGAAAGCUCCCUAGAGCAAAGCUUUUCACAAAACAAAAAGCUCGCGGCAG---------- .......(((((......(((........(((((((....))))))).(((....)))...))).((((....))))..(((((((.........))))))).)))))..---------- ( -31.00) >DroSec_CAF1 10419 105 + 1 AUUAAACGACCAAUCCCGUUUAUCAAUAUA-UUUUGGCGCCAAAAAAUCGCGU--------GUAAGCUC------GAAAAGCUUUUGGAAUGCGAAGAAGCUUUCACCGAGCUUAGCAAC ..((((((........))))))........-(((((....)))))....((..--------.(((((((------(.(((((((((.........)))))))))...))))))))))... ( -26.70) >DroSim_CAF1 12855 105 + 1 AUCAAACGACCAAUCCCGUUUAUCAAUAUA-UUUUGGCGCCAAAAAAUCGCGU--------GUAAGCUC------GAAAAGCUUUUGGAAUGCGAAGAAGCUUUCGCCGAGCUUAGCAAC ...(((((........))))).........-(((((....)))))....((..--------.(((((((------(.(((((((((.........)))))))))...))))))))))... ( -25.60) >DroEre_CAF1 32329 105 + 1 ACCAAACGACCAAUCCCGUUUAUCAAUGUA-UUUUGGCGCCAAAAAAUCGUGC--------GUAAGCUC------GGAAAGCUUUUCGAUUACGAAGAAGCUUUCGCCGAGCUUAGCAAC ...(((((........))))).........-(((((....))))).....(((--------.(((((((------((.((((((((((....)).))))))))...)))))))))))).. ( -33.20) >DroYak_CAF1 13505 105 + 1 AUCAAACGACCAAUCCCGUUUAUCAAUACA-UUUUGGCGCCAAAAAACCGUGC--------GUAAGCUC------GGAAAGCUUUUCGAAUGCGAAAAAGCUUUCGCCGAGCUUAGUAAC ...(((((........))))).....(((.-.....((((.........))))--------.(((((((------((.((((((((((....)).))))))))...)))))))))))).. ( -31.70) >consensus AUCAAACGACCAAUCCCGUUUAUCAAUAUA_UUUUGGCGCCAAAAAAUCGCGC________GUAAGCUC______GGAAAGCUUUUCGAAUGCGAAGAAGCUUUCGCCGAGCUUAGCAAC ...(((((........)))))...............((((.........)))).........(((((((......((((.((((((.........))))))))))...)))))))..... (-14.56 = -14.95 + 0.39)


| Location | 10,164,470 – 10,164,572 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -25.58 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10164470 102 + 22224390 CAAAAAAUCGCGCGUAAGCUCGGAAAGCUUUUCGAAUGCGAAGAAGCUUUCGCCAAGCUUAGCAACAACUGCAGCGGAACGAAAUGGCAAUGGGAACAGGCC .......((.(((.((((((.((.((((((((((....)).))))))))...)).))))))(((.....))).))))).......(((..((....)).))) ( -30.70) >DroSec_CAF1 10458 102 + 1 CAAAAAAUCGCGUGUAAGCUCGAAAAGCUUUUGGAAUGCGAAGAAGCUUUCACCGAGCUUAGCAACAACUGCAGCGGAACGAAAUGGCAAUGGAAACACGAC .......((((...((((((((.(((((((((.........)))))))))...))))))))(((.....))).)))).............((....)).... ( -28.60) >DroSim_CAF1 12894 102 + 1 CAAAAAAUCGCGUGUAAGCUCGAAAAGCUUUUGGAAUGCGAAGAAGCUUUCGCCGAGCUUAGCAACAACUGCAGCGGAACGAAAUGGCAAUGGAAACACGAC .......((((...((((((((.(((((((((.........)))))))))...))))))))(((.....))).)))).............((....)).... ( -28.60) >DroEre_CAF1 32368 102 + 1 CAAAAAAUCGUGCGUAAGCUCGGAAAGCUUUUCGAUUACGAAGAAGCUUUCGCCGAGCUUAGCAACAACUGCAGCGGAACGAAAUGGCAAUGGGAACAAGCC .......((((.(((((((((((.((((((((((....)).))))))))...)))))))).(((.....))).)))..))))...(((..((....)).))) ( -35.50) >DroYak_CAF1 13544 102 + 1 CAAAAAACCGUGCGUAAGCUCGGAAAGCUUUUCGAAUGCGAAAAAGCUUUCGCCGAGCUUAGUAACAACCGCAGCGGAACGAAAUGGAAAUGAAAAGAAGCC .......((((((((((((((((.((((((((((....)).))))))))...)))))))))((....)))))).)))......................... ( -32.00) >consensus CAAAAAAUCGCGCGUAAGCUCGGAAAGCUUUUCGAAUGCGAAGAAGCUUUCGCCGAGCUUAGCAACAACUGCAGCGGAACGAAAUGGCAAUGGAAACAAGCC .......((((...((((((((((((((((((((....)).)))))))))..)))))))))(((.....))).)))).............((....)).... (-25.58 = -26.26 + 0.68)


| Location | 10,164,470 – 10,164,572 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
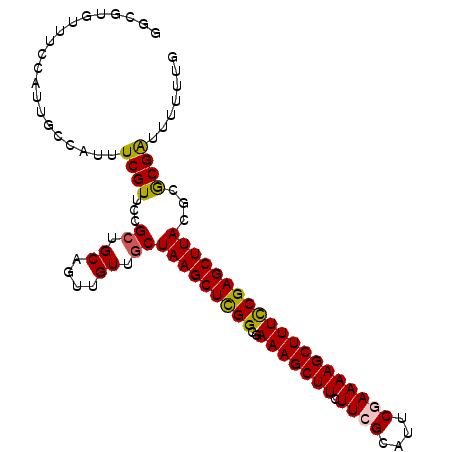
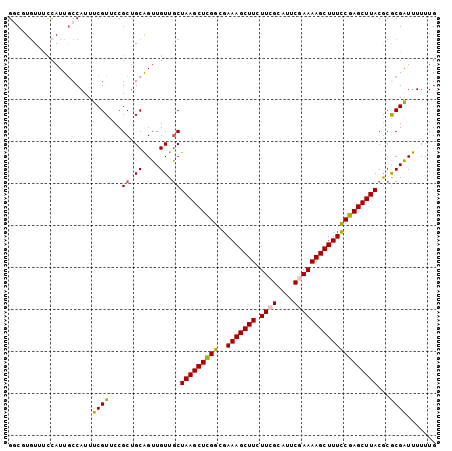
>X_DroMel_CAF1 10164470 102 - 22224390 GGCCUGUUCCCAUUGCCAUUUCGUUCCGCUGCAGUUGUUGCUAAGCUUGGCGAAAGCUUCUUCGCAUUCGAAAAGCUUUCCGAGCUUACGCGCGAUUUUUUG (((.((....))..)))...............((((((.(((((((((((..(((((((.((((....)))))))))))))))))))).))))))))..... ( -29.80) >DroSec_CAF1 10458 102 - 1 GUCGUGUUUCCAUUGCCAUUUCGUUCCGCUGCAGUUGUUGCUAAGCUCGGUGAAAGCUUCUUCGCAUUCCAAAAGCUUUUCGAGCUUACACGCGAUUUUUUG (((((((....................((.((....)).))((((((((..((((((((.............)))))))))))))))).)))))))...... ( -26.12) >DroSim_CAF1 12894 102 - 1 GUCGUGUUUCCAUUGCCAUUUCGUUCCGCUGCAGUUGUUGCUAAGCUCGGCGAAAGCUUCUUCGCAUUCCAAAAGCUUUUCGAGCUUACACGCGAUUUUUUG (((((((....................((.((....)).))((((((((..((((((((.............)))))))))))))))).)))))))...... ( -26.12) >DroEre_CAF1 32368 102 - 1 GGCUUGUUCCCAUUGCCAUUUCGUUCCGCUGCAGUUGUUGCUAAGCUCGGCGAAAGCUUCUUCGUAAUCGAAAAGCUUUCCGAGCUUACGCACGAUUUUUUG (((.((....))..)))...............((((((.(((((((((((..(((((((.((((....)))))))))))))))))))).))))))))..... ( -34.30) >DroYak_CAF1 13544 102 - 1 GGCUUCUUUUCAUUUCCAUUUCGUUCCGCUGCGGUUGUUACUAAGCUCGGCGAAAGCUUUUUCGCAUUCGAAAAGCUUUCCGAGCUUACGCACGGUUUUUUG .........................(((.((((((....))(((((((((..(((((((((.((....)))))))))))))))))))))))))))....... ( -31.20) >consensus GGCGUGUUUCCAUUGCCAUUUCGUUCCGCUGCAGUUGUUGCUAAGCUCGGCGAAAGCUUCUUCGCAUUCGAAAAGCUUUCCGAGCUUACGCGCGAUUUUUUG ....................((((...((.((....)).))(((((((((..(((((((.((((....))))))))))))))))))))...))))....... (-24.00 = -23.80 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:41 2006