
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,163,717 – 10,163,867 |
| Length | 150 |
| Max. P | 0.938194 |

| Location | 10,163,717 – 10,163,836 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.40 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
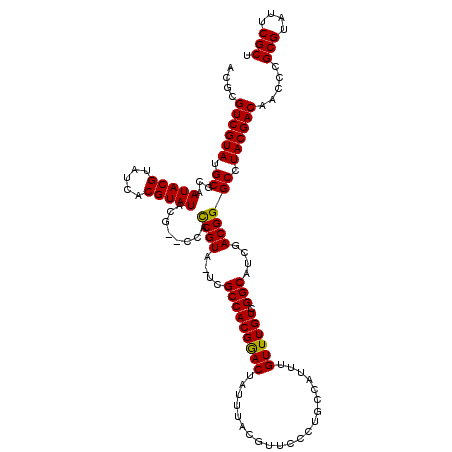
>X_DroMel_CAF1 10163717 119 - 22224390 ACGCGUCGUAUGCGCAAUACGUAUCACGUAUACGUACCAUCGUA-UCGCCACGGACUAUUUACGUUCCCUGCCAUUUGUUUGUCGGCAUCGACGGGCCUACGACAACCCGCGUAUUCGCC ....((((((.((....(((((((.....)))))))((.(((.(-(.(((((((((.....................)))))).)))))))).)))).)))))).....(((....))). ( -34.00) >DroSec_CAF1 9757 117 - 1 ACGCGUCGUAUGCGCAAUACGUAUCACGUAUACG--CCACCGUA-UCGCCACGGACUAUUUACGUUCCCAGCCAUUUGUUUGUCGGCAUCGACGGGCCUACGACAACCCGCGUAUUCGCU ....((((((.((((.(((((.....)))))..)--)..((((.-..(((((((((.....................)))))).)))....)))))).)))))).....(((....))). ( -34.60) >DroSim_CAF1 12162 117 - 1 ACGCGUCGUAUGCGCAAUACGUAUCACGUAUACG--CCACCGUA-UCGCCACGGACUAUUUACGUUCCCAGCCAUUUGUUUGUCGGCAUCGACGGGCCUACGACAACCCGCGUAUUCGCU ....((((((.((((.(((((.....)))))..)--)..((((.-..(((((((((.....................)))))).)))....)))))).)))))).....(((....))). ( -34.60) >DroEre_CAF1 31536 118 - 1 ACGCGUCGUAUGCGCAAUACGUAUCACGUAUACG--CCACCGUAAUCGCCACGAACUAUUUACGUUCUCUGCCAUUUGUUUGUCGGCAUCGACGGGCCUACGACAACCCGCGUAUUCGCU ....((((((.((((.(((((.....)))))..)--)..((((....(((((((((.....................)))))).)))....)))))).)))))).....(((....))). ( -35.40) >DroYak_CAF1 12687 117 - 1 ACGCGUCGUAUGCGCAAUACGUAUCACGUAUACG--CCACCGUA-UCGCCACGGACUAUUUACGUUCUCUGCCAUUUGUUUGUCGGCAUCGACGGGCCUACGACAACCCGCGUAUUCGCU ....((((((.((((.(((((.....)))))..)--)..((((.-..(((((((((.....................)))))).)))....)))))).)))))).....(((....))). ( -34.60) >consensus ACGCGUCGUAUGCGCAAUACGUAUCACGUAUACG__CCACCGUA_UCGCCACGGACUAUUUACGUUCCCUGCCAUUUGUUUGUCGGCAUCGACGGGCCUACGACAACCCGCGUAUUCGCU ....((((((.((...(((((.....)))))........((((....(((((((((.....................)))))).)))....)))))).)))))).....(((....))). (-33.72 = -33.40 + -0.32)

| Location | 10,163,757 – 10,163,867 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 97.07 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -28.18 |
| Energy contribution | -28.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
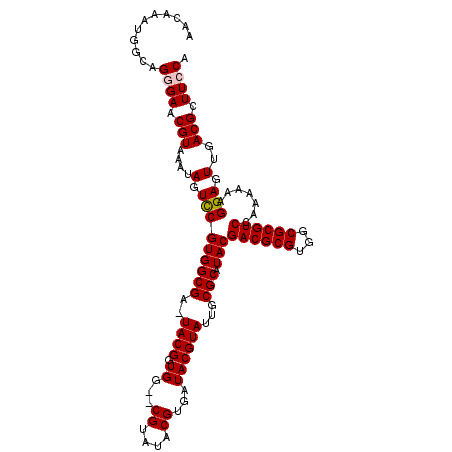
>X_DroMel_CAF1 10163757 110 + 22224390 AACAAAUGGCAGGGAACGUAAAUAGUCCGUGGCGA-UACGAUGGUACGUAUACGUGAUACGUAUUGCGCAUACGACGCGUGGCGCGUCCAAAAAAGGAGUUGACGCUUCCA ...........((((.(((....(.((((((((((-((((.((.((((....)))).)))))))))).)))..((((((...)))))).......))).)..))).)))). ( -36.70) >DroSec_CAF1 9797 108 + 1 AACAAAUGGCUGGGAACGUAAAUAGUCCGUGGCGA-UACGGUGG--CGUAUACGUGAUACGUAUUGCGCAUACGACGCGUGGCGCGUCCAAAAAAGGAGUUGACGCUUCCA ............(((((((....(.((((((((((-((((((.(--((....))).)).)))))))).)))..((((((...)))))).......))).)..))).)))). ( -37.60) >DroSim_CAF1 12202 108 + 1 AACAAAUGGCUGGGAACGUAAAUAGUCCGUGGCGA-UACGGUGG--CGUAUACGUGAUACGUAUUGCGCAUACGACGCGUGGCGCGUCCAAAAAAGGAGUUGACGCUUCCA ............(((((((....(.((((((((((-((((((.(--((....))).)).)))))))).)))..((((((...)))))).......))).)..))).)))). ( -37.60) >DroEre_CAF1 31576 109 + 1 AACAAAUGGCAGAGAACGUAAAUAGUUCGUGGCGAUUACGGUGG--CGUAUACGUGAUACGUAUUGCGCAUACGACGCGUGGCGCGUCCAAAAAAGGAGUUGACGCUUCCA ....(((.((...((((.......))))...)).)))..((.((--((((((((.....))))).((((.((((...)))))))).(((......)))....))))).)). ( -33.30) >DroYak_CAF1 12727 108 + 1 AACAAAUGGCAGAGAACGUAAAUAGUCCGUGGCGA-UACGGUGG--CGUAUACGUGAUACGUAUUGCGCAUACGACGCGUGGCGCGUCCAAAAAAGGAGUUGACGCUUCCA ...........(.((((((....(.((((((((((-((((((.(--((....))).)).)))))))).)))..((((((...)))))).......))).)..))).)))). ( -34.00) >consensus AACAAAUGGCAGGGAACGUAAAUAGUCCGUGGCGA_UACGGUGG__CGUAUACGUGAUACGUAUUGCGCAUACGACGCGUGGCGCGUCCAAAAAAGGAGUUGACGCUUCCA ...........((((.(((....(.(((((((((..((((.((...((....))...))))))...))).)))((((((...)))))).......))).)..))).)))). (-28.18 = -28.42 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:39 2006