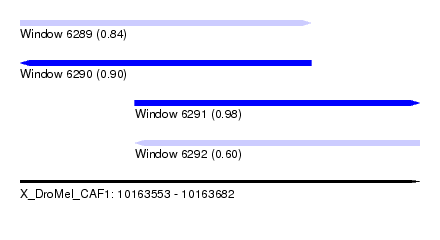
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,163,553 – 10,163,682 |
| Length | 129 |
| Max. P | 0.984742 |

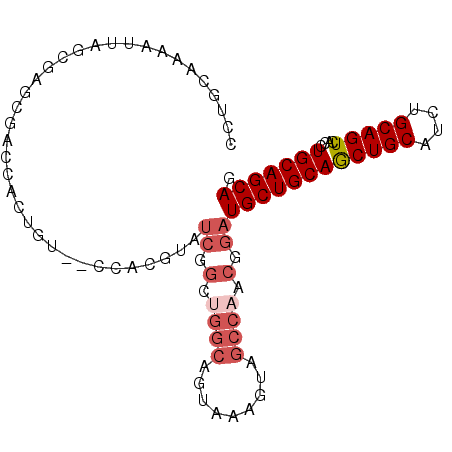
| Location | 10,163,553 – 10,163,647 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -19.20 |
| Energy contribution | -20.64 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10163553 94 + 22224390 CCUGCAAAAUUAGCGAGCGACCACUGU--CCACGUAUCGGCUGGCAGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAG .((((.......((.(((....((((.--(((.((....)))))))))...(((((........))))).)))))...(((((....))))))))) ( -30.90) >DroSec_CAF1 9591 96 + 1 CGUGCAAAAUUAGCGAGCAACCACUGUCCCCACGUAUCGGCUGGCAGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAG (((((.......))(.((.((.((((((.((.......))..))))))...)).))).)))..((((((((((((....))))....)))))))). ( -30.30) >DroSim_CAF1 11996 96 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAG ...............................................................((((((((((((....))))....)))))))). ( -18.10) >DroEre_CAF1 31350 94 + 1 CCUUUGAAAUUAGCGAGCGACCACUGU--CCACGUAUCGGCUGGCUGUAAAGUAGCCAACGGAUGCUGCAACUGCAUCUGCAGUCACCUGCAGCAG ............(((.(.(((....))--)).)))(((.(.((((((.....)))))).).)))(((((((((((....)))))....)))))).. ( -31.40) >DroYak_CAF1 12511 94 + 1 CCUUAAAAAUUAGCGAACGACCACUGU--CCAAAUAUCGGUGGGCUGUAACGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAG ((...................(((((.--........)))))(((((.....)))))...)).((((((((((((....))))....)))))))). ( -30.20) >consensus CCUGCAAAAUUAGCGAGCGACCACUGU__CCACGUAUCGGCUGGCAGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAG ....................................((.(.((((.........)))).).))((((((((((((....)))))....))))))). (-19.20 = -20.64 + 1.44)


| Location | 10,163,553 – 10,163,647 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -22.27 |
| Energy contribution | -24.24 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10163553 94 - 22224390 CUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACUGCCAGCCGAUACGUGG--ACAGUGGUCGCUCGCUAAUUUUGCAGG ...((((((....)))))).(((((..((((((((.((((((.........)))))).))).....(--((....)))))).)).....))))).. ( -35.80) >DroSec_CAF1 9591 96 - 1 CUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACUGCCAGCCGAUACGUGGGGACAGUGGUUGCUCGCUAAUUUUGCACG ...((((((....))))))..(((((..(.(.(((.((((((.........)))))).))).).((....)))..)))))..((.......))... ( -39.70) >DroSim_CAF1 11996 96 - 1 CUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .((((((((....((((....))))))))))))............................................................... ( -17.70) >DroEre_CAF1 31350 94 - 1 CUGCUGCAGGUGACUGCAGAUGCAGUUGCAGCAUCCGUUGGCUACUUUACAGCCAGCCGAUACGUGG--ACAGUGGUCGCUCGCUAAUUUCAAAGG ...((((((....)))))).((.((((((((((((.(((((((.......))))))).))).....(--((....)))))).)).)))).)).... ( -34.40) >DroYak_CAF1 12511 94 - 1 CUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACGUUACAGCCCACCGAUAUUUGG--ACAGUGGUCGUUCGCUAAUUUUUAAGG .((((((((....((((....))))))))))))...((((((.(((((((.(.(((........)))--.).)))).)))..))))))........ ( -29.40) >consensus CUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACAGCCAGCCGAUACGUGG__ACAGUGGUCGCUCGCUAAUUUUGAAGG ..(((((((....((((....)))))))))))(((.((((((.........)))))).)))..........(((((....)))))........... (-22.27 = -24.24 + 1.97)

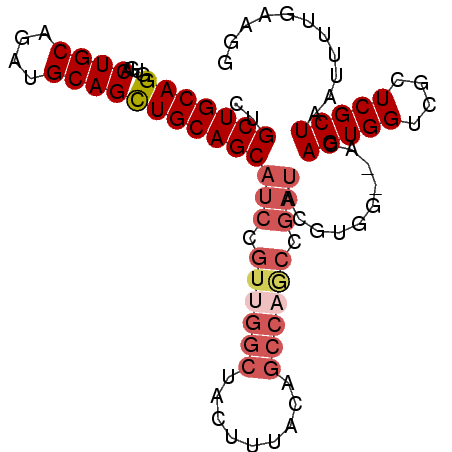
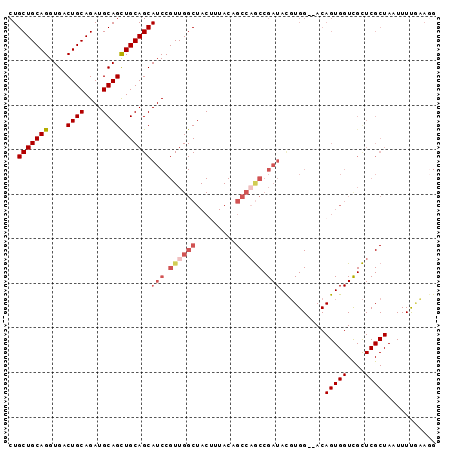
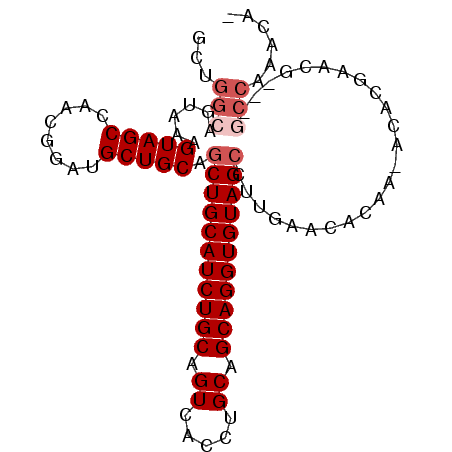
| Location | 10,163,590 – 10,163,682 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -24.60 |
| Energy contribution | -25.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10163590 92 + 22224390 GCUGGCAGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAGGUGUAGCCUUGAACACAA-ACACAAACG---GCCAAAUA- ..((((.((...(((((........))))).(((((((((((.((.....)).)))))))))))...........-......)).---))))....- ( -31.40) >DroSec_CAF1 9630 92 + 1 GCUGGCAGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAGGUGUAGCCUUGAACACAA-ACACGAACG---GCCAAACA- ..((((.((...(((((........))))).(((((((((((.((.....)).)))))))))))...........-......)).---))))....- ( -31.40) >DroSim_CAF1 12035 92 + 1 NNNNNNNGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAGGUGUAGCCUUGAACACAA-ACACGAACG---GCCAAACA- ...............(((......((((((((((((....))))....)))))))).((((........))))..-........)---))......- ( -26.30) >DroEre_CAF1 31387 96 + 1 GCUGGCUGUAAAGUAGCCAACGGAUGCUGCAACUGCAUCUGCAGUCACCUGCAGCAGGUGUAGCCUUGAACACAA-ACACGAACGGCGGCCAAACAG ..(((((((...((..........((((((((((((....)))))....))))))).((((........))))..-......)).)))))))..... ( -32.40) >DroYak_CAF1 12548 93 + 1 GUGGGCUGUAACGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAGGUGUAGCCUUGAACACAAAACACGAAUG---UCCAAACA- ((.(((((.....))))).))((((((((((.((((...(((((....))))))))).)))))).(((....))).........)---))).....- ( -33.10) >consensus GCUGGCAGUAAAGUAGCCAACGGAUGCUGCAGCUGCAUCUGCAGUCACCUGCAGCAGGUGUAGCCUUGAACACAA_ACACGAACG___GCCAAACA_ ...(((......(((((........))))).(((((((((((.((.....)).)))))))))))........................)))...... (-24.60 = -25.60 + 1.00)

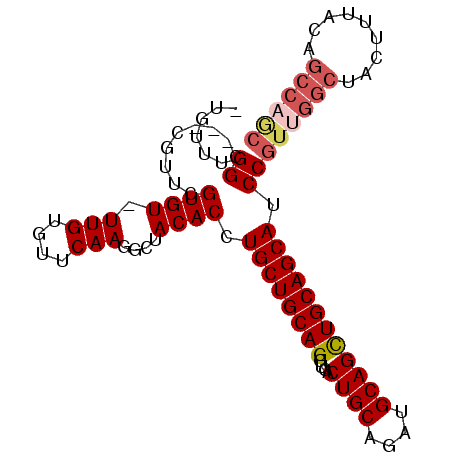
| Location | 10,163,590 – 10,163,682 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -24.36 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10163590 92 - 22224390 -UAUUUGGC---CGUUUGUGU-UUGUGUUCAAGGCUACACCUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACUGCCAGC -....((((---(.....((.-.......)).)))))....((((((((....((((....))))))))))))...((((((.........)))))) ( -31.00) >DroSec_CAF1 9630 92 - 1 -UGUUUGGC---CGUUCGUGU-UUGUGUUCAAGGCUACACCUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACUGCCAGC -.((.((((---(.....((.-.......)).))))).)).((((((((....((((....))))))))))))...((((((.........)))))) ( -32.10) >DroSim_CAF1 12035 92 - 1 -UGUUUGGC---CGUUCGUGU-UUGUGUUCAAGGCUACACCUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACNNNNNNN -....((((---((..((.(.-..(((((...((((.((....((((((....)))))).)).))))..)))))))).))))))............. ( -28.90) >DroEre_CAF1 31387 96 - 1 CUGUUUGGCCGCCGUUCGUGU-UUGUGUUCAAGGCUACACCUGCUGCAGGUGACUGCAGAUGCAGUUGCAGCAUCCGUUGGCUACUUUACAGCCAGC ...((((..(((.........-..)))..))))(((.((.(((((((((....)))))...)))).)).)))....(((((((.......))))))) ( -32.90) >DroYak_CAF1 12548 93 - 1 -UGUUUGGA---CAUUCGUGUUUUGUGUUCAAGGCUACACCUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACGUUACAGCCCAC -..((((((---(((.........)))))))))(((.((.(((((((((....)))))...)))).)).)))....((.((((.......)))).)) ( -32.10) >consensus _UGUUUGGC___CGUUCGUGU_UUGUGUUCAAGGCUACACCUGCUGCAGGUGACUGCAGAUGCAGCUGCAGCAUCCGUUGGCUACUUUACAGCCAGC ......((.........((((.(((....)))....)))).((((((((....((((....)))))))))))).))((((((.........)))))) (-24.36 = -25.48 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:37 2006