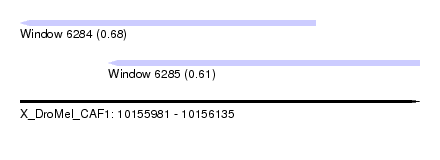
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,155,981 – 10,156,135 |
| Length | 154 |
| Max. P | 0.676640 |

| Location | 10,155,981 – 10,156,095 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -15.22 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10155981 114 - 22224390 ACAAAAGACGUAGUUUCAUUAAUUGAAAUAAAUAGCGAAAAAAAAAGUACUGAAGGAGCUGCUGUUUUCUUGUUGAACGAAACAAAAGAAAAAGUAUUAAGCAGCCCAAGACGA------ ........(((.((((((.....)))))).........................((.((((((.((((((((((......))))..)))))).......))))))))...))).------ ( -23.80) >DroSec_CAF1 2431 110 - 1 ACAAAAGAAGCAGCUUCAUUAAUUGAAAUAAAUAGCGAAA---GAAGCACUGA-GGAGCUGCUGUUUUGUUGUUGAACGAAACAAAAGAAAAAGAAUUAAGCAGCCAAAGACGA------ ........(((((((((....(((......))).((....---...)).....-)))))))))((((((((....))))))))...............................------ ( -22.50) >DroSim_CAF1 5003 110 - 1 ACAAAAGACGCAGCUUCAUUAAUUGAAAUAAAUAGCGAAA---GAAGCACUGA-GGAGCUGCUGUUUUGUUGUUGAACGAAACAAAAGAAAAAGAAUUCGGCAGCCAAAGACGA------ .........((((((((....(((......))).((....---...)).....-))))))))(((((((((((((((...................)))))))))..)))))).------ ( -24.11) >DroEre_CAF1 24130 110 - 1 ACAAAAGACGUAGCUUCAUUAAUUGAAAUAAAUAGCGAAA---AAAGCACUGA-GGAGCUGCUAUUUUCUUGUUGAACGAAACAAAAGGAAAAGAAUUAGACGGCCUAAGACGA------ .........((((((((....(((......))).((....---...)).....-))))))))..(((((((.(((.......))).)))))))(..((((.....))))..)..------ ( -18.90) >DroYak_CAF1 4800 116 - 1 CCAAAAGACGCAGCUUCAUUAAUUGAAAUAAAUAGGGAAA---AAAGCACUGA-GGAGCUGCUAUUCCGUUGUUAGACGAAACAAAAGAAAAAGAAUUAAGCAGCCUUAGACGAAGACGA ........((...((((....(((......))).......---......((((-(..((((((((((((((....))))...(....).....))))..)))))))))))..)))).)). ( -19.10) >consensus ACAAAAGACGCAGCUUCAUUAAUUGAAAUAAAUAGCGAAA___AAAGCACUGA_GGAGCUGCUGUUUUGUUGUUGAACGAAACAAAAGAAAAAGAAUUAAGCAGCCAAAGACGA______ .........((((((((....(((......))).((..........))......))))))))..((((.(((((......))))).)))).............................. (-15.22 = -14.74 + -0.48)

| Location | 10,156,015 – 10,156,135 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.84 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10156015 120 - 22224390 AUGUGUGUUGAGAGCCCUUUUGUUUGGGAUUAAUUAAGCGACAAAAGACGUAGUUUCAUUAAUUGAAAUAAAUAGCGAAAAAAAAAGUACUGAAGGAGCUGCUGUUUUCUUGUUGAACGA ..((..(((((...(((........))).)))))...))((((((((((((((((((....(((......))).((..........))......)))))))).))))).)))))...... ( -22.40) >DroSec_CAF1 2465 116 - 1 AAGUGUGUUGAGAGCCCUUUUCUUUGGGAUUAAUUAAGCGACAAAAGAAGCAGCUUCAUUAAUUGAAAUAAAUAGCGAAA---GAAGCACUGA-GGAGCUGCUGUUUUGUUGUUGAACGA ......(((((...(((........))).)))))..((((((((((..(((((((((....(((......))).((....---...)).....-))))))))).))))))))))...... ( -32.60) >DroSim_CAF1 5037 116 - 1 AAGUGUGUUGAGAGCCCUUUUGUUUGGGAUUAAUUAGGCGACAAAAGACGCAGCUUCAUUAAUUGAAAUAAAUAGCGAAA---GAAGCACUGA-GGAGCUGCUGUUUUGUUGUUGAACGA ......(((((...(((........))).)))))..((((((((((.(.((((((((....(((......))).((....---...)).....-))))))))).))))))))))...... ( -31.40) >DroEre_CAF1 24164 116 - 1 AAGUGUGUUGAGAGCCCUUUUGUUUGGGAUUAAUUAAGCGACAAAAGACGUAGCUUCAUUAAUUGAAAUAAAUAGCGAAA---AAAGCACUGA-GGAGCUGCUAUUUUCUUGUUGAACGA ..((..(((((...(((........))).)))))...))(((((.(((.((((((((....(((......))).((....---...)).....-))))))))...))).)))))...... ( -24.00) >DroYak_CAF1 4840 116 - 1 AAGUGUGUUGAGAGCCCUUUUGUUUGGGAUUAAUUAAGCGCCAAAAGACGCAGCUUCAUUAAUUGAAAUAAAUAGGGAAA---AAAGCACUGA-GGAGCUGCUAUUCCGUUGUUAGACGA ....(((((.....(((((((((((.(.((((((.((((((........)).)))).))))))).))))))).))))...---..)))))...-((((......))))(((....))).. ( -28.20) >consensus AAGUGUGUUGAGAGCCCUUUUGUUUGGGAUUAAUUAAGCGACAAAAGACGCAGCUUCAUUAAUUGAAAUAAAUAGCGAAA___AAAGCACUGA_GGAGCUGCUGUUUUGUUGUUGAACGA ...((((((((...(((........))).)))))..((((((((((.(.((((((((....(((......))).((..........))......))))))))).))))))))))..))). (-23.36 = -23.84 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:31 2006