
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,146,580 – 10,146,750 |
| Length | 170 |
| Max. P | 0.969831 |

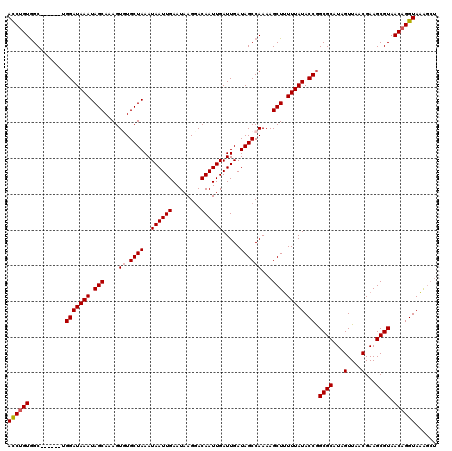
| Location | 10,146,580 – 10,146,694 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.06 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10146580 114 + 22224390 ACCUGUGGC------UGGAUAAAUAGCAAAGUGUGCUAAAUAAUUGAAUAAGGACAAUUGAUUGAUAGCCAAAAGCUUUUUAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCU ((((((.((------((((((((.(((....((.((((..((((((........)))))).....))))))...))).))))).)))))((...(....)...))...))))))...... ( -30.00) >DroSec_CAF1 35803 114 + 1 ACCUGUGGC------UGGAUAAAUAGCAAAGUCUGCUAAAUAAUUGAAUAAGGACAAUUGAUUGAUAGCCAAAAGCUUUUUAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCU ((((((.((------((((((((.(((...(.(((.(((.((((((........)))))).))).))).)....))).))))).)))))((...(....)...))...))))))...... ( -27.50) >DroSim_CAF1 28211 114 + 1 ACCUGUGGC------UGGAUAAAUAGCAAAGUCUGCUAAAUAAUUGAAUAAGGACAAUUGAUUGAUAGCCAAAAGCUUUUUAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCU ((((((.((------((((((((.(((...(.(((.(((.((((((........)))))).))).))).)....))).))))).)))))((...(....)...))...))))))...... ( -27.50) >DroYak_CAF1 28843 114 + 1 ACCUGUGGC------UGGAUAAAUAGCAAAGUGUGCUAAAUAAUUGAAUAAGUACAAUUGAUUGAUAGCCAAAAGCUCUUUAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAGAGCU ((((((.((------((((((((.(((....((.((((..((((((........)))))).....))))))...))).))))).)))))((...(....)...))...))))))...... ( -29.80) >DroAna_CAF1 50243 118 + 1 AUCCGUAGCCAGGUGAGGAUAAAUAGCAAAGUGUGCUAAAUAAUUGAAUAAGGACAAUUGAUUGAUAGCCAAAAGCUUUUUAUACCAGCGCAUAGUUAACGAAGCGUAACAGGUAAGA-- .......(((......(((((((.(((....((.((((..((((((........)))))).....))))))...))).))))).)).((((...(....)...))))....)))....-- ( -22.30) >consensus ACCUGUGGC______UGGAUAAAUAGCAAAGUGUGCUAAAUAAUUGAAUAAGGACAAUUGAUUGAUAGCCAAAAGCUUUUUAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCU ((((((..........(((((((.(((....((.((((..((((((........)))))).....))))))...))).))))).)).((((...(....)...)))).))))))...... (-22.62 = -23.06 + 0.44)


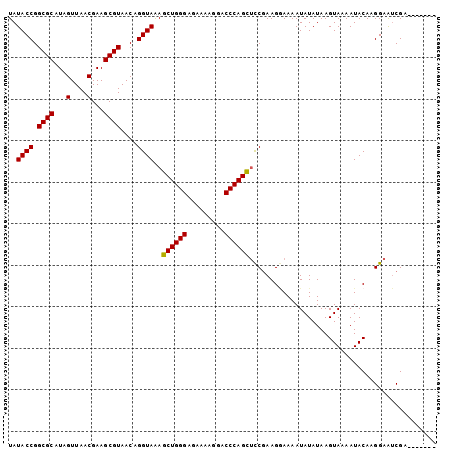
| Location | 10,146,580 – 10,146,694 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -22.56 |
| Consensus MFE | -20.84 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10146580 114 - 22224390 AGCUUUACCUGUUACGCUUCGUUAACUAUGCGCCGGUAUAAAAAGCUUUUGGCUAUCAAUCAAUUGUCCUUAUUCAAUUAUUUAGCACACUUUGCUAUUUAUCCA------GCCACAGGU ......((((((...((............))((.((.(((((.(((...((((((..(((.(((((........)))))))))))).))....))).))))))).------)).)))))) ( -23.90) >DroSec_CAF1 35803 114 - 1 AGCUUUACCUGUUACGCUUCGUUAACUAUGCGCCGGUAUAAAAAGCUUUUGGCUAUCAAUCAAUUGUCCUUAUUCAAUUAUUUAGCAGACUUUGCUAUUUAUCCA------GCCACAGGU ......((((((...((............))((.((.(((((.(((.....((((..(((.(((((........)))))))))))).......))).))))))).------)).)))))) ( -23.10) >DroSim_CAF1 28211 114 - 1 AGCUUUACCUGUUACGCUUCGUUAACUAUGCGCCGGUAUAAAAAGCUUUUGGCUAUCAAUCAAUUGUCCUUAUUCAAUUAUUUAGCAGACUUUGCUAUUUAUCCA------GCCACAGGU ......((((((...((............))((.((.(((((.(((.....((((..(((.(((((........)))))))))))).......))).))))))).------)).)))))) ( -23.10) >DroYak_CAF1 28843 114 - 1 AGCUCUACCUGUUACGCUUCGUUAACUAUGCGCCGGUAUAAAGAGCUUUUGGCUAUCAAUCAAUUGUACUUAUUCAAUUAUUUAGCACACUUUGCUAUUUAUCCA------GCCACAGGU ......((((((...((............))((.((.(((((.(((...((((((..(((.(((((........)))))))))))).))....))).))))))).------)).)))))) ( -23.90) >DroAna_CAF1 50243 118 - 1 --UCUUACCUGUUACGCUUCGUUAACUAUGCGCUGGUAUAAAAAGCUUUUGGCUAUCAAUCAAUUGUCCUUAUUCAAUUAUUUAGCACACUUUGCUAUUUAUCCUCACCUGGCUACGGAU --......((((...(((.(((.......)))..((.(((((.(((...((((((..(((.(((((........)))))))))))).))....))).)))))))......))).)))).. ( -18.80) >consensus AGCUUUACCUGUUACGCUUCGUUAACUAUGCGCCGGUAUAAAAAGCUUUUGGCUAUCAAUCAAUUGUCCUUAUUCAAUUAUUUAGCACACUUUGCUAUUUAUCCA______GCCACAGGU ......((((((..(((............)))..((.(((((.(((.....((((..(((.(((((........)))))))))))).......))).)))))))..........)))))) (-20.84 = -20.88 + 0.04)

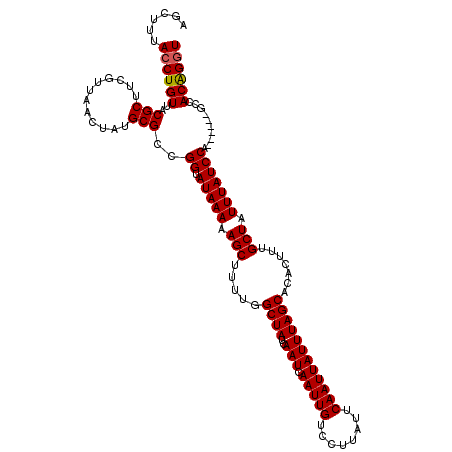

| Location | 10,146,654 – 10,146,750 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.61 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10146654 96 + 22224390 UAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCUGGGUGAAAAGAACCCAGUUCCGAAGCAAAAUAUAUAAGUAAAAUACAAGGAGACGA------- ..((((.((((...(....)...))))....)))).((((((((.......)))))))).............................(....)..------- ( -22.50) >DroSec_CAF1 35877 96 + 1 UAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCUGGGAGAAAAGGACCCAGCUCCGAAGGAAAAUAUAUAAGUAAAAUACAAGGAAUCGA------- ..((((.((((...(....)...))))....))))..((((((.........))))))(((...........................))).....------- ( -20.33) >DroSim_CAF1 28285 96 + 1 UAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCUGGGAGAAAAGGACCCAGCUCCAAAGGAAAAUAUAUAAGUAAAAUACAAGGAAUCGA------- ..((((.((((...(....)...))))....)))).(((((((.........))))))).....................................------- ( -20.20) >DroYak_CAF1 28917 103 + 1 UAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAGAGCUGGGAGUAGAGGACCCAGCAUCGAAGAAAAAUAUAUAAGUAAAAUACAAGGAAUCGGAAUCAGA ..((((.((((...(....)...))))....))))((((((((.........)))))).)).......................................... ( -20.80) >consensus UAUACCGGCGCAUAGUUAACGAAGCGUAACAGGUAAAGCUGGGAGAAAAGGACCCAGCUCCGAAGGAAAAUAUAUAAGUAAAAUACAAGGAAUCGA_______ ..((((.((((...(....)...))))....))))..((((((.........))))))............................................. (-19.51 = -19.32 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:29 2006