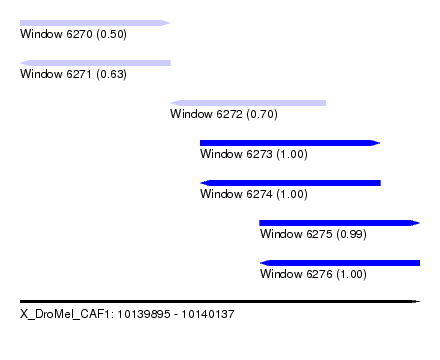
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,139,895 – 10,140,137 |
| Length | 242 |
| Max. P | 0.996767 |

| Location | 10,139,895 – 10,139,986 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.74 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -16.35 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10139895 91 + 22224390 GUGUGUGUGCCUGCUGCAGCGCACAUUACACACGCACACUCGUACAUUAAAUUUAUGUGCUCGAGUUGGACGCGGUA-AAAAAAAAAGAGUA (((((((((((((...))).))))).))))).(((.(((((((((((.......))))))..))).))...)))...-.............. ( -26.60) >DroSec_CAF1 29322 83 + 1 ---UGUGUGCCUGCUGCAG--CACAUUACACACGCACACUCAUACAUUAAAUUUAUGUGCUCGAGUUGGACGCG----AACAAAAAAGAGUA ---.((((.((.(((..((--(((((............................)))))))..))).)))))).----.............. ( -16.69) >DroSim_CAF1 20771 86 + 1 GUAUGUGUGCCUGCUGGAG--CACAUUACACACGCACACUCGUACAUUAAAUUUAUGUGCUCGAGUUGGACGCG----AACAAAAAAGAGUA ((.(((((.((.(((.(((--(((((............................)))))))).))).)))))))----.))........... ( -24.69) >DroYak_CAF1 21587 90 + 1 GUGUGUGUGUCUGCUGCAG--CACAUUACACACGCACACUCGUACAUUAAAUUUAUGUGCUCGAGUUGGACGCCAAAAAAAAAAAAAGAGUA ((((((((((.(((....)--))....))))))))))((((((((((.......)))))).....((((...))))...........)))). ( -28.50) >consensus GUGUGUGUGCCUGCUGCAG__CACAUUACACACGCACACUCGUACAUUAAAUUUAUGUGCUCGAGUUGGACGCG____AAAAAAAAAGAGUA (((((((((..((.((.....))))...)))))))))((((((((((.......))))))((......)).................)))). (-16.35 = -17.60 + 1.25)


| Location | 10,139,895 – 10,139,986 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 90.74 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.45 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10139895 91 - 22224390 UACUCUUUUUUUUU-UACCGCGUCCAACUCGAGCACAUAAAUUUAAUGUACGAGUGUGCGUGUGUAAUGUGCGCUGCAGCAGGCACACACAC .............(-(((((((..((.((((.(((...........))).))))))..)))).))))(((((.(((...))))))))..... ( -24.90) >DroSec_CAF1 29322 83 - 1 UACUCUUUUUUGUU----CGCGUCCAACUCGAGCACAUAAAUUUAAUGUAUGAGUGUGCGUGUGUAAUGUG--CUGCAGCAGGCACACA--- ..........((((----((.(.....).))))))..................((((((.((((((.....--.)))).)).)))))).--- ( -22.60) >DroSim_CAF1 20771 86 - 1 UACUCUUUUUUGUU----CGCGUCCAACUCGAGCACAUAAAUUUAAUGUACGAGUGUGCGUGUGUAAUGUG--CUCCAGCAGGCACACAUAC (((((......(((----((.(.....).)))))((((.......))))..)))))...((((((....((--(....))).)))))).... ( -22.20) >DroYak_CAF1 21587 90 - 1 UACUCUUUUUUUUUUUUUGGCGUCCAACUCGAGCACAUAAAUUUAAUGUACGAGUGUGCGUGUGUAAUGUG--CUGCAGCAGACACACACAC .................(((...)))(((((.(((...........))).)))))(((.((((((....((--(....))).)))))).))) ( -20.30) >consensus UACUCUUUUUUGUU____CGCGUCCAACUCGAGCACAUAAAUUUAAUGUACGAGUGUGCGUGUGUAAUGUG__CUGCAGCAGGCACACACAC ...................(((....(((((.(((...........))).))))).)))((((((..(((........))).)))))).... (-18.83 = -18.45 + -0.38)



| Location | 10,139,986 – 10,140,080 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10139986 94 - 22224390 AGGCGUGGCCCGCAAAGUGGGCAUAAUGUUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU-UCAUUUUCUUUUCAUUUUGUGCUCUU ((((((.((((((...))))))...))))))((..(....((....((((((((..............-...)))))))).))....)..))... ( -20.33) >DroSec_CAF1 29405 94 - 1 AGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU-UCAUUUUCCUUUCAUUUUGUGCUCUU ((((((.((((((...))))))...))))))((..(((((......))))(((((.............-.....)))))........)..))... ( -22.17) >DroSim_CAF1 20857 94 - 1 AGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU-UCAUUUUCCUUUCAUUUUGUGCUCUU ((((((.((((((...))))))...))))))((..(((((......))))(((((.............-.....)))))........)..))... ( -22.17) >DroYak_CAF1 21677 95 - 1 AGGCGUGGCCCGCAAAGUGGGCAUAAUGUUUAGUGCCUUUUGUUGUAAAGAGUCUUUAUAUUUUUUUUUACAUUUUCUUUUCAUUUUGUGCUCUG .(((...)))((((((((((((((........)))))......(((((((((...........))))))))).........)))))))))..... ( -22.40) >consensus AGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU_UCAUUUUCCUUUCAUUUUGUGCUCUU ((((((.((((((...))))))...))))))....((((((......)))))).......................................... (-19.08 = -19.08 + -0.00)



| Location | 10,140,004 – 10,140,113 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -33.24 |
| Energy contribution | -33.55 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10140004 109 + 22224390 GAAAAUGA-AAAUAAAAUAAGACUCCUCUUUACAACAAAAGGCACUAAACAUUAUGCCCACUUUGCGGGCCACGCCUACCAAGGGGGCGUGGCGAAAAAAGGGGCUG-------CUG ........-..........((...(((((((.(..((((.((((..........))))...))))..)(((((((((.(....))))))))))....)))))))...-------)). ( -32.70) >DroSec_CAF1 29423 109 + 1 GAAAAUGA-AAAUAAAAUAAGACUCCUCUUUACAACAAAAGGCACUAAGCAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCGGAAAAAGGGGCUG-------CUG ........-.................(((((......))))).....((((....((((.((((....(((((((((((...)))))))))))....))))))))))-------)). ( -37.40) >DroSim_CAF1 20875 109 + 1 GAAAAUGA-AAAUAAAAUAAGACUCCUCUUUACAACAAAAGGCACUAAGCAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCGGAAAAAGGGGCUG-------CUG ........-.................(((((......))))).....((((....((((.((((....(((((((((((...)))))))))))....))))))))))-------)). ( -37.40) >DroYak_CAF1 21695 117 + 1 GAAAAUGUAAAAAAAAAUAUAAAGACUCUUUACAACAAAAGGCACUAAACAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCAAAAAAAGAGGCUGAUGGCUGCCG .....((((((.................))))))......((((.....(((((.(((..((((....(((((((((((...)))))))))))....)))).))))))))..)))). ( -41.63) >consensus GAAAAUGA_AAAUAAAAUAAGACUCCUCUUUACAACAAAAGGCACUAAACAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCGAAAAAAGGGGCUG_______CUG ........................(((((((.(..((((.((((..........))))...))))..)(((((((((((...)))))))))))....)))))))............. (-33.24 = -33.55 + 0.31)



| Location | 10,140,004 – 10,140,113 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -34.38 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10140004 109 - 22224390 CAG-------CAGCCCCUUUUUUCGCCACGCCCCCUUGGUAGGCGUGGCCCGCAAAGUGGGCAUAAUGUUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU-UCAUUUUC ...-------..((.(((((((..((((((((((...))..))))))))..((((...((((((........))))))....))))))))))).))............-........ ( -33.30) >DroSec_CAF1 29423 109 - 1 CAG-------CAGCCCCUUUUUCCGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU-UCAUUUUC .((-------((((((((((....((((((((((....)).))))))))....)))).))))....)))).((..((((((......))))))...))..........-........ ( -39.60) >DroSim_CAF1 20875 109 - 1 CAG-------CAGCCCCUUUUUCCGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU-UCAUUUUC .((-------((((((((((....((((((((((....)).))))))))....)))).))))....)))).((..((((((......))))))...))..........-........ ( -39.60) >DroYak_CAF1 21695 117 - 1 CGGCAGCCAUCAGCCUCUUUUUUUGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGUUUAGUGCCUUUUGUUGUAAAGAGUCUUUAUAUUUUUUUUUACAUUUUC .(((........))).........((((((((((....)).))))))))..(((((..((((((........)))))))))))(((((((((...........)))))))))..... ( -39.50) >consensus CAG_______CAGCCCCUUUUUCCGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUUUGUUGUAAAGAGGAGUCUUAUUUUAUUU_UCAUUUUC ...............(((((((..((((((((((....)).))))))))..((((...((((((........))))))....)))))))))))........................ (-34.38 = -34.88 + 0.50)

| Location | 10,140,040 – 10,140,137 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -35.44 |
| Energy contribution | -35.94 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10140040 97 + 22224390 AAAGGCACUAAACAUUAUGCCCACUUUGCGGGCCACGCCUACCAAGGGGGCGUGGCGAAAAAAGGGGCUG-------CUGCCUGCCAUUAACAUGAAGAAGAAA ...((((.....((.((.((((.((((....(((((((((.(....))))))))))....))))))))))-------.))..)))).................. ( -34.70) >DroSec_CAF1 29459 95 + 1 AAAGGCACUAAGCAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCGGAAAAAGGGGCUG-------CUGCCUGCCAUUAAUAUGAA--AGAAA ...((((...((((....((((.((((....(((((((((((...)))))))))))....))))))))))-------))...))))...........--..... ( -44.70) >DroSim_CAF1 20911 95 + 1 AAAGGCACUAAGCAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCGGAAAAAGGGGCUG-------CUGCCUGCCAUUAACAUGAA--AGAAA ...((((...((((....((((.((((....(((((((((((...)))))))))))....))))))))))-------))...))))...........--..... ( -44.70) >DroYak_CAF1 21732 97 + 1 AAAGGCACUAAACAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCAAAAAAAGAGGCUGAUGGCUGCCGGCUGCCAUCAACAUGAA------- ...((((.....(((((.(((..((((....(((((((((((...)))))))))))....)))).))))))))((.....))))))...........------- ( -41.60) >consensus AAAGGCACUAAACAUUAUGCCCACUUUGCGGGCCACGCCUCCCAAGGGGGCGUGGCGAAAAAAGGGGCUG_______CUGCCUGCCAUUAACAUGAA__AGAAA ...((((...........((((.((((....(((((((((((...)))))))))))....))))))))..............)))).................. (-35.44 = -35.94 + 0.50)

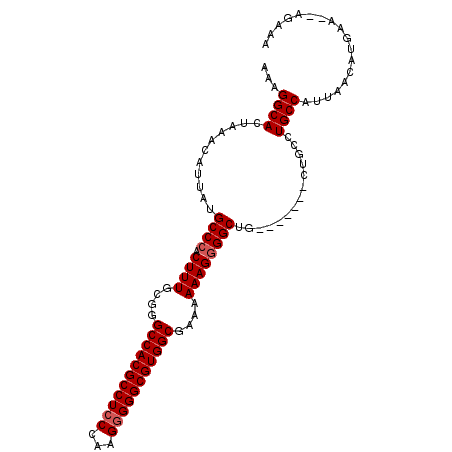
| Location | 10,140,040 – 10,140,137 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -33.97 |
| Energy contribution | -33.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10140040 97 - 22224390 UUUCUUCUUCAUGUUAAUGGCAGGCAG-------CAGCCCCUUUUUUCGCCACGCCCCCUUGGUAGGCGUGGCCCGCAAAGUGGGCAUAAUGUUUAGUGCCUUU ..................((((((((.-------..((((((((....((((((((((...))..))))))))....)))).))))....))))...))))... ( -33.90) >DroSec_CAF1 29459 95 - 1 UUUCU--UUCAUAUUAAUGGCAGGCAG-------CAGCCCCUUUUUCCGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUU .....--...........((((((((.-------..((((((((....((((((((((....)).))))))))....)))).))))....))))...))))... ( -41.70) >DroSim_CAF1 20911 95 - 1 UUUCU--UUCAUGUUAAUGGCAGGCAG-------CAGCCCCUUUUUCCGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUU .....--...........((((((((.-------..((((((((....((((((((((....)).))))))))....)))).))))....))))...))))... ( -41.70) >DroYak_CAF1 21732 97 - 1 -------UUCAUGUUGAUGGCAGCCGGCAGCCAUCAGCCUCUUUUUUUGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGUUUAGUGCCUUU -------.....(((((((((........)))))))))(.((((....((((((((((....)).))))))))....)))).)(((((........)))))... ( -43.50) >consensus UUUCU__UUCAUGUUAAUGGCAGGCAG_______CAGCCCCUUUUUCCGCCACGCCCCCUUGGGAGGCGUGGCCCGCAAAGUGGGCAUAAUGCUUAGUGCCUUU ..................((((((((..........((((((((....((((((((((....)).))))))))....)))).))))....))))...))))... (-33.97 = -33.97 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:23 2006