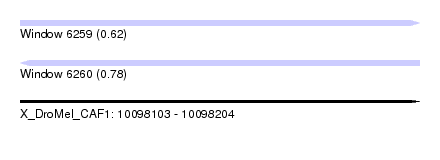
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,098,103 – 10,098,204 |
| Length | 101 |
| Max. P | 0.780522 |

| Location | 10,098,103 – 10,098,204 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -18.97 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10098103 101 + 22224390 UGCGAUGCACCUA-------GCCCACUGCAC-GUGGGCGCCAUCUGU-GGCAUGCAAAUGCAACUGCCUGCUGUAUUACAUGGCGUAUACUUGAUAUGCCUCGCUUUUUA .((((((((....-------((((((.....-))))))((((....)-))).))))((((((.(.....).))))))....(((((((.....)))))))))))...... ( -37.80) >DroSec_CAF1 16118 101 + 1 UGUGAUGCACCUC-------GCCCACUGCAC-GUGGGCGCCAUCUGU-AGCAUGCAAAUGCAACUGCCUGCUGUAUUACAUGGCGUAUACUUGAUAUGCCUCGCUUUUUA ((((((((....(-------((((((.....-)))))))........-((((.(((........))).)))))))))))).(((((((.....))))))).......... ( -36.80) >DroSim_CAF1 15817 101 + 1 UGUGAUGCACCUC-------GCCCAGUGCAC-GUGGGCGCCAUCUGU-AGCAUGCAAAUGCAACUGCCUGCUGUAUUACAUGGCGUAUACUUGAUAUGCCUCGCUUUUUA ((((((((....(-------((((((....)-.))))))........-((((.(((........))).)))))))))))).(((((((.....))))))).......... ( -33.40) >DroEre_CAF1 20389 101 + 1 UGUUAUGCAGCCC-------ACCCAUUGCAC-GCGGGCGCCAUCUGU-GGCAUGCAAAUGCAACUGCUUGCUGGACUACAUGGCGAAUACUUGAUAUGCCUCGCUUUUUA .((((((.((.((-------(......(((.-((((..((((....)-))).(((....))).)))).)))))).)).)))))).......................... ( -26.60) >DroYak_CAF1 13581 101 + 1 UGUAAUGCACCUC-------GACCACUGCAC-GUGGGCGCCAUCUGU-GGCAUGCAAGUGCAACUGCCUGCUGGAUUACACGGCGUAUACUUGAUAUGCCUGGCUUUUUA (((((((((....-------......)))..-(..(((((((....)-))).(((....)))...)))..)...)))))).(((((((.....))))))).......... ( -30.70) >DroAna_CAF1 26819 100 + 1 CCCAAUGCAUUUACAUUCUGCAUAAGUGCAUGGCUGGCGCCAUCUUUCAGCAUGCAACCGCAACUGCAAUCUGGGCCACAUGGCGUAUACGUGAU----------UUUUG .(((.((((((((.........)))))))))))...(((((((..(((((..((((........))))..)))))....))))))).........----------..... ( -26.10) >consensus UGUGAUGCACCUC_______GCCCACUGCAC_GUGGGCGCCAUCUGU_AGCAUGCAAAUGCAACUGCCUGCUGGAUUACAUGGCGUAUACUUGAUAUGCCUCGCUUUUUA ....((((............((((((......))))))(((((.((..((((.(((........))).))))....)).)))))))))...................... (-18.97 = -20.38 + 1.42)

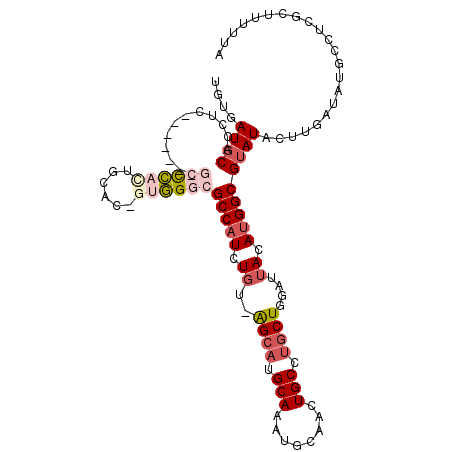
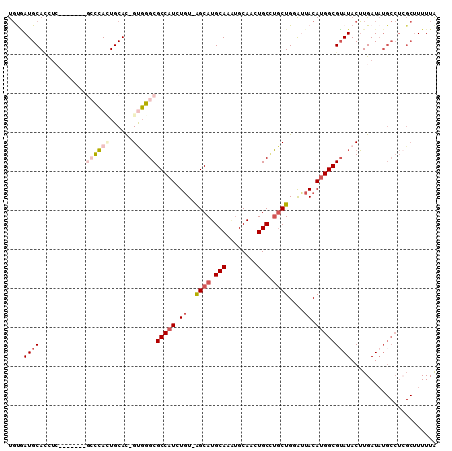
| Location | 10,098,103 – 10,098,204 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -19.39 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10098103 101 - 22224390 UAAAAAGCGAGGCAUAUCAAGUAUACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCC-ACAGAUGGCGCCCAC-GUGCAGUGGGC-------UAGGUGCAUCGCA ......((((.((((...........(((((((......)).((((..(((....)))))))-....)))))((((((-.....))))))-------...)))).)))). ( -40.10) >DroSec_CAF1 16118 101 - 1 UAAAAAGCGAGGCAUAUCAAGUAUACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCU-ACAGAUGGCGCCCAC-GUGCAGUGGGC-------GAGGUGCAUCACA ......(.((.((((.((........(((((.(.....((((.((((......)))).))))-..).)))))((((((-.....))))))-------)).)))).)).). ( -37.00) >DroSim_CAF1 15817 101 - 1 UAAAAAGCGAGGCAUAUCAAGUAUACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCU-ACAGAUGGCGCCCAC-GUGCACUGGGC-------GAGGUGCAUCACA ......(.((.((((.((........(((((.(.....((((.((((......)))).))))-..).)))))(((((.-(....))))))-------)).)))).)).). ( -32.80) >DroEre_CAF1 20389 101 - 1 UAAAAAGCGAGGCAUAUCAAGUAUUCGCCAUGUAGUCCAGCAAGCAGUUGCAUUUGCAUGCC-ACAGAUGGCGCCCGC-GUGCAAUGGGU-------GGGCUGCAUAACA ......(((((((.......)).))))).((((((((((.(.....(((((((.((..((((-(....)))))..)).-))))))).).)-------))))))))).... ( -35.90) >DroYak_CAF1 13581 101 - 1 UAAAAAGCCAGGCAUAUCAAGUAUACGCCGUGUAAUCCAGCAGGCAGUUGCACUUGCAUGCC-ACAGAUGGCGCCCAC-GUGCAGUGGUC-------GAGGUGCAUUACA ..........(((.(((.....))).))).((((((.((.(.(((..((((((.((..((((-(....)))))..)).-))))))..)))-------..).)).)))))) ( -30.70) >DroAna_CAF1 26819 100 - 1 CAAAA----------AUCACGUAUACGCCAUGUGGCCCAGAUUGCAGUUGCGGUUGCAUGCUGAAAGAUGGCGCCAGCCAUGCACUUAUGCAGAAUGUAAAUGCAUUGGG .....----------.((((((.......))))))(((((..((((.(((((.(((((((.((....(((((....))))).))..)))))))..))))).))))))))) ( -29.80) >consensus UAAAAAGCGAGGCAUAUCAAGUAUACGCCAUGUAAUACAGCAGGCAGUUGCAUUUGCAUGCC_ACAGAUGGCGCCCAC_GUGCAGUGGGC_______GAGGUGCAUCACA ....................((((..(((((.(......(((.((((......)))).)))....).)))))(((((........)))))..........))))...... (-19.39 = -19.70 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:09 2006