
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,094,895 – 10,094,992 |
| Length | 97 |
| Max. P | 0.894508 |

| Location | 10,094,895 – 10,094,992 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -18.32 |
| Energy contribution | -19.29 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10094895 97 - 22224390 UAGCAAGUAUUUUGUUGCGUUUGCCGUUAGCAGUACCGUUACAUGUGCUGGCU----CAUGUAAAUAUUGCCGUUAC-------A-------UACGGCAGC----CAUAUUGUUGAUGC- ..(((.(((......)))...)))(((((((((((...(((((((.(....).----)))))))...(((((((...-------.-------.))))))).----..))))))))))).- ( -30.20) >DroSec_CAF1 11232 97 - 1 UAGCAACUAUAUUGUUGCGUUUGCCUUUAGCAGUACCGUUACAUGUGCUGGCU----CGUGUAAACAUUACCGUUAC-------A-------UACGGCAGC----CAUAUUGUUGAUGC- ...((((.((((.(((((((((((....((((((((........))))).)))----...)))))).....(((...-------.-------.))))))))----.)))).))))....- ( -27.80) >DroSim_CAF1 11652 97 - 1 UAGCAACUAUAUUGUUGCGUUUGCCUUUGGCAGUACCGUUACAUGUGCUGGCU----CGUGUAAACAUUACCGUUAC-------A-------UACGGCAGC----CAUAUUGUUGAUGC- ...((((.((((.(((((((((((....(.((((((........)))))).).----...)))))).....(((...-------.-------.))))))))----.)))).))))....- ( -30.00) >DroEre_CAF1 17164 104 - 1 UAGCAACUAUUUUGUUGCGUUUGCCGUUAGCAGUACCGUUACAUGUGCUGGCG----UGUAUAAACAUUGCCGUUAC-------AUACGCCACACGGCAGC----CAUAUUGUUGAUGC- ..(((((......)))))......(((((((((((..(((...((((.(((((----(((...(((......)))..-------))))))))))))..)))----..))))))))))).- ( -35.30) >DroYak_CAF1 10228 111 - 1 UAGCAACUAUUUUGUUGCGUUUGCCGUUAGCAGUACCGUUACAUGUGCUGGCG----UGUGUAAACAUUGCCGUUACUUACGACAUACGACAUACGGCAGC----CAUAUUGUUGAUGC- ..(((((......)))))......(((((((((((........((.((((.((----(((((......((.(((.....))).))....))))))).))))----))))))))))))).- ( -36.70) >DroAna_CAF1 21032 99 - 1 UGGCAGCCGAAG-GUCGGG--GGCUGU----UGUACCAUUACAUGUGCUGGCUGUCGUGGGUAAACAUUGCCGUUAC-------A-------UACGGCAACGGUGCGUAUGCGUUAUGCG .(((((((....-......--))))))----)(((((........((((.((....)).))))....(((((((...-------.-------.))))))).)))))(((((...))))). ( -33.50) >consensus UAGCAACUAUAUUGUUGCGUUUGCCGUUAGCAGUACCGUUACAUGUGCUGGCU____CGUGUAAACAUUGCCGUUAC_______A_______UACGGCAGC____CAUAUUGUUGAUGC_ ..(((((......)))))......(((((((((((...(((((((............)))))))...(((((((...................))))))).......))))))))))).. (-18.32 = -19.29 + 0.98)

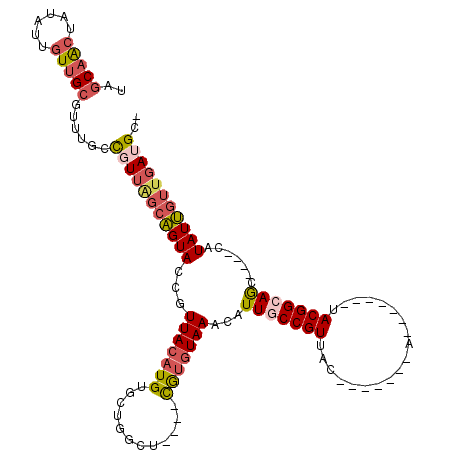
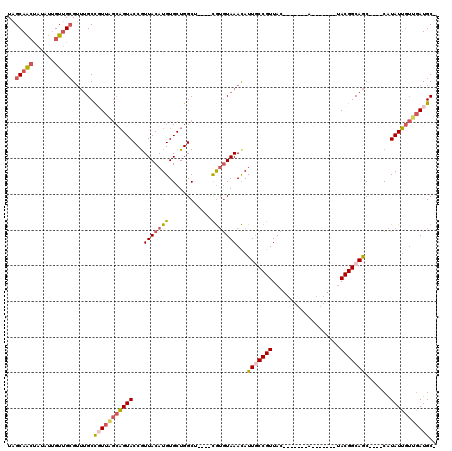
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:05 2006