
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,088,175 – 10,088,399 |
| Length | 224 |
| Max. P | 0.999999 |

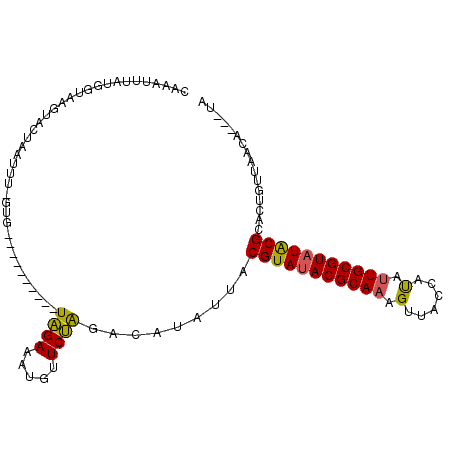
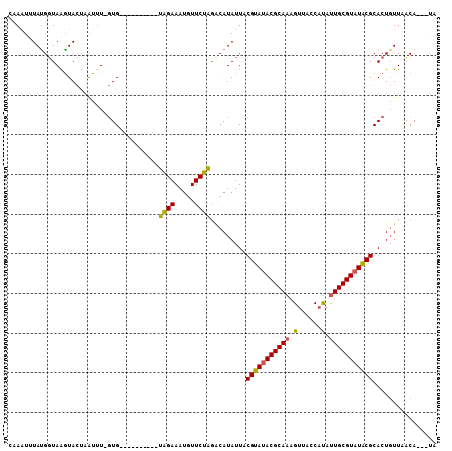
| Location | 10,088,175 – 10,088,272 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.17 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 5.14 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088175 97 + 22224390 CUAAUUUAUGGUAGGUAUCAAUUU-UUG----------UAGAAAUUUUCUGUUCAUAUUACGUAUACGCAAAGUUAGCACAUUGCGUAUACGCACUUUUUACAUUUUA ........((((....))))....-.((----------((((((......((.......))((((((((((.((....)).))))))))))....))))))))..... ( -19.00) >DroEre_CAF1 10550 95 + 1 UAAAUUUACGUUAAGUACUAAUAUAGUG----------UGGAAAUGUUCCAGGCAUAUUGCGUAUACGCAAAGGUACCAUGCUGCGUAUACGCACUGUUCACA---AA .........(...(((...(((((.((.----------((((.....)))).)))))))(((((((((((..(......)..))))))))))))))...)...---.. ( -27.20) >DroYak_CAF1 3286 105 + 1 CCAUUGUAUGGUAAAUACUUGUUUUGUGCAAUUAUUUCUAGAAAUGUUCUAGACAUAUUACGUAUACGCAAAGUAAUCAUAUUGCGUUUGCGCACUGUUAUCA---UA ......((((((((...........((((...(((.((((((.....)))))).)))....(((.((((((.((....)).)))))).)))))))..))))))---)) ( -25.12) >consensus CAAAUUUAUGGUAAGUACUAAUUU_GUG__________UAGAAAUGUUCUAGACAUAUUACGUAUACGCAAAGUUACCAUAUUGCGUAUACGCACUGUUAACA___UA ......................................((((.....)))).........(((((((((((.(......).)))))))))))................ (-14.90 = -14.47 + -0.44)

| Location | 10,088,175 – 10,088,272 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.17 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -16.88 |
| Energy contribution | -16.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.76 |
| SVM decision value | 6.58 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088175 97 - 22224390 UAAAAUGUAAAAAGUGCGUAUACGCAAUGUGCUAACUUUGCGUAUACGUAAUAUGAACAGAAAAUUUCUA----------CAA-AAAUUGAUACCUACCAUAAAUUAG .....(((......(((((((((((((.((....)).)))))))))))))......)))...........----------...-........................ ( -21.60) >DroEre_CAF1 10550 95 - 1 UU---UGUGAACAGUGCGUAUACGCAGCAUGGUACCUUUGCGUAUACGCAAUAUGCCUGGAACAUUUCCA----------CACUAUAUUAGUACUUAACGUAAAUUUA ..---.(((..((.(((((((((((((...(....).)))))))))))))...))..((((.....))))----------)))......................... ( -25.30) >DroYak_CAF1 3286 105 - 1 UA---UGAUAACAGUGCGCAAACGCAAUAUGAUUACUUUGCGUAUACGUAAUAUGUCUAGAACAUUUCUAGAAAUAAUUGCACAAAACAAGUAUUUACCAUACAAUGG ..---........((((....((((((..........))))))........(((.((((((.....)))))).)))...)))).......((((.....))))..... ( -20.10) >consensus UA___UGUAAACAGUGCGUAUACGCAAUAUGAUAACUUUGCGUAUACGUAAUAUGACUAGAACAUUUCUA__________CAC_AAAUUAGUACUUACCAUAAAUUAG ..............(((((((((((((..........)))))))))))))........(((.....)))....................................... (-16.88 = -16.67 + -0.22)

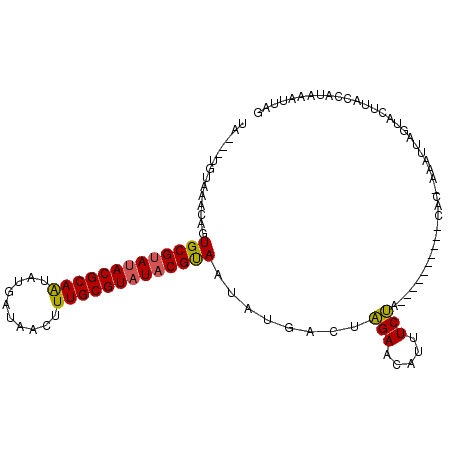
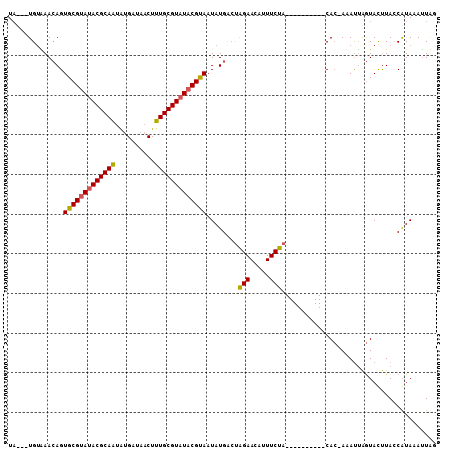
| Location | 10,088,202 – 10,088,298 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -17.56 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.69 |
| SVM decision value | 6.73 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088202 96 + 22224390 ----------UAGAAAUUUUCUGUUCAUAUUACGUAUACGCAAAGUUAGCACAUUGCGUAUACGCACUUUUUACAUUUUAUUGCGUAUACGCACUAUUUUCACACU ----------..((((.....(((........(((((((((((.((....)).)))))))))))........)))......((((....))))....))))..... ( -22.59) >DroEre_CAF1 10578 93 + 1 ----------UGGAAAUGUUCCAGGCAUAUUGCGUAUACGCAAAGGUACCAUGCUGCGUAUACGCACUGUUCACA---AAUUGCGUAUACGCGCUGUUAUCCUAUC ----------((((.....))))......(((((....)))))(((((.((.((.(((((((((((.((....))---...))))))))))))))).)).)))... ( -31.50) >DroYak_CAF1 3314 103 + 1 CAAUUAUUUCUAGAAAUGUUCUAGACAUAUUACGUAUACGCAAAGUAAUCAUAUUGCGUUUGCGCACUGUUAUCA---UACGACGUAUACGCACUGUUAUCAAACU ....(((.((((((.....)))))).)))...((((((((....(((((.(((.((((....)))).))).))..---)))..))))))))............... ( -22.20) >consensus __________UAGAAAUGUUCUAGACAUAUUACGUAUACGCAAAGUUACCAUAUUGCGUAUACGCACUGUUAACA___UAUUGCGUAUACGCACUGUUAUCAAACU ..........((((.....)))).........(((((((((((.(....)....((((....))))..............)))))))))))............... (-17.56 = -17.90 + 0.34)



| Location | 10,088,202 – 10,088,298 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -21.95 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 6.71 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088202 96 - 22224390 AGUGUGAAAAUAGUGCGUAUACGCAAUAAAAUGUAAAAAGUGCGUAUACGCAAUGUGCUAACUUUGCGUAUACGUAAUAUGAACAGAAAAUUUCUA---------- ..(((....(((.(((((((((((((......(((.....((((....))))...))).....))))))))))))).)))..)))...........---------- ( -28.90) >DroEre_CAF1 10578 93 - 1 GAUAGGAUAACAGCGCGUAUACGCAAUU---UGUGAACAGUGCGUAUACGCAGCAUGGUACCUUUGCGUAUACGCAAUAUGCCUGGAACAUUUCCA---------- ....(((.....(((((((((((((...---((....)).))))))))))).))..((((...(((((....)))))..)))).........))).---------- ( -36.00) >DroYak_CAF1 3314 103 - 1 AGUUUGAUAACAGUGCGUAUACGUCGUA---UGAUAACAGUGCGCAAACGCAAUAUGAUUACUUUGCGUAUACGUAAUAUGUCUAGAACAUUUCUAGAAAUAAUUG .(((....)))..(((((((((((.(..---((((.....((((....)))).....))))..).))))))))))).(((.((((((.....)))))).))).... ( -27.90) >consensus AGUAUGAUAACAGUGCGUAUACGCAAUA___UGUAAACAGUGCGUAUACGCAAUAUGAUAACUUUGCGUAUACGUAAUAUGACUAGAACAUUUCUA__________ ..............((((((((((((..............((((....))))...........)))))))))))).........(((.....)))........... (-21.95 = -21.18 + -0.77)


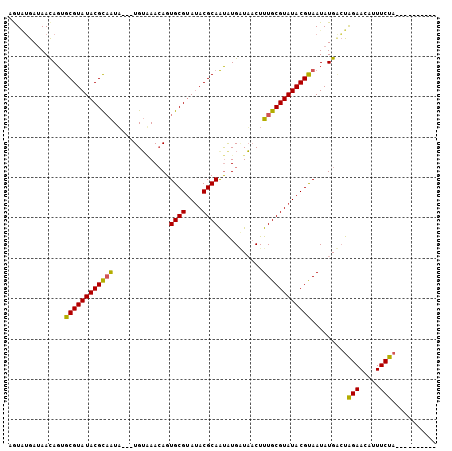
| Location | 10,088,232 – 10,088,337 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -24.03 |
| Energy contribution | -23.93 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.80 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088232 105 + 22224390 CAAAGUUAGCACAUUGCGUAUACGCACUUUUUACAUUUUAUUGCGUAUACGCACUAUUUUCACACUACGUAUACGCACUGUUAUGGCAUUACGUAUACGCCCUGU ..........(((..(((((((((.........(((..((.((((((((((................)))))))))).))..)))......)))))))))..))) ( -29.45) >DroEre_CAF1 10608 102 + 1 CAAAGGUACCAUGCUGCGUAUACGCACUGUUCACA---AAUUGCGUAUACGCGCUGUUAUCCUAUCACGUAUACGCACUGUUGUCACAUUGCGUAUACGCACUGU ...(((((.((.((.(((((((((((.((....))---...))))))))))))))).)).)))....((((((((((.(((....))).))))))))))...... ( -39.40) >DroYak_CAF1 3354 102 + 1 CAAAGUAAUCAUAUUGCGUUUGCGCACUGUUAUCA---UACGACGUAUACGCACUGUUAUCAAACUACGUAUGCGCACUGUUAUCAUACGACGUAUACGCACUGU ..........(((.(((((.(((((...((.....---.))(.((((((((....(((....)))..))))))))).............).)))).))))).))) ( -21.50) >consensus CAAAGUUACCAUAUUGCGUAUACGCACUGUUAACA___UAUUGCGUAUACGCACUGUUAUCAAACUACGUAUACGCACUGUUAUCACAUUACGUAUACGCACUGU ..............((((((((((((...............))))))))))))..............((((((((...(((....)))...))))))))...... (-24.03 = -23.93 + -0.10)


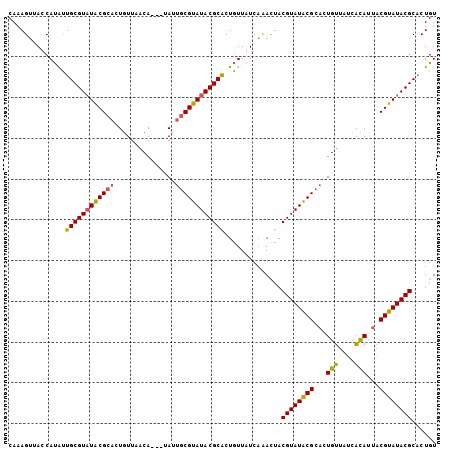
| Location | 10,088,232 – 10,088,337 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -28.31 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -5.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088232 105 - 22224390 ACAGGGCGUAUACGUAAUGCCAUAACAGUGCGUAUACGUAGUGUGAAAAUAGUGCGUAUACGCAAUAAAAUGUAAAAAGUGCGUAUACGCAAUGUGCUAACUUUG (((..(((((((((((....(((.....((((((((((((.(((....))).)))))))))))).....))).......)))))))))))..))).......... ( -43.20) >DroEre_CAF1 10608 102 - 1 ACAGUGCGUAUACGCAAUGUGACAACAGUGCGUAUACGUGAUAGGAUAACAGCGCGUAUACGCAAUU---UGUGAACAGUGCGUAUACGCAGCAUGGUACCUUUG .((.((((((((((((.(((.((((...((((((((((((...(.....)..))))))))))))..)---)))..))).))))))))))))...))......... ( -45.60) >DroYak_CAF1 3354 102 - 1 ACAGUGCGUAUACGUCGUAUGAUAACAGUGCGCAUACGUAGUUUGAUAACAGUGCGUAUACGUCGUA---UGAUAACAGUGCGCAAACGCAAUAUGAUUACUUUG ....(((((((..((((((((((....((((((((...((......))...))))))))..))))))---))))....))))))).................... ( -27.90) >consensus ACAGUGCGUAUACGUAAUGUGAUAACAGUGCGUAUACGUAGUAUGAUAACAGUGCGUAUACGCAAUA___UGUAAACAGUGCGUAUACGCAAUAUGAUAACUUUG .....(((((((((((............((((((((((((.(........).))))))))))))...............)))))))))))............... (-28.31 = -29.10 + 0.78)



| Location | 10,088,272 – 10,088,374 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 61.22 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -11.01 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088272 102 + 22224390 UUGCGUAUACGCACUAUUUUCACACUACGUAUACGCACUGUUAUGGCAUUACGUAUACGCCCUGUAUUCUAAUUACGAAAAGUUGACUUUUGGCUCAUUUUA- .((((((((((................))))))))))..((((((......)))).))(((.((((.......))))(((((....))))))))........- ( -21.39) >DroSec_CAF1 4635 102 + 1 UUGCGUAUACGCAAUGUAAUUAUAAUACGUAUACGCACUGUUACAAUAUUACGUAUACGCCCUGUAUUCUGAUUACGAAGAGUUGCUUUUUCGCUCUUUUUA- (((((....))))).(((((((.((((((((((((...(((....)))...))))))).....))))).)))))))(((((((.........)))))))...- ( -24.30) >DroSim_CAF1 4080 78 + 1 UUGCGUAUACGGACUGUUACAAUGUUACGUAUACGCCCUGU------AUU------------------CUGAUUACGAAGAGUUGCUUUUUGGCUCUUUUUA- ..((((((((((((.........))).)))))))))...((------(..------------------.....)))((((((((.......))))))))...- ( -20.80) >DroEre_CAF1 10645 81 + 1 UUGCGUAUACGCGCUGUUAUCCUAUCACGUAUACGCACUGUUGUCACAUUGCGUAUACGCACUGUGUUCAUAUUAC---------------------UUUUA- .((((((((((.(.((......)).).))))))))))....((.((((.((((....)))).))))..))......---------------------.....- ( -23.90) >DroYak_CAF1 3391 82 + 1 CGACGUAUACGCACUGUUAUCAAACUACGUAUGCGCACUGUUAUCAUACGACGUAUACGCACUGUUAUCAAACUAC---------------------GUAAAU .(.((((((((....(((....)))..((((((...........)))))).)))))))))................---------------------...... ( -14.60) >consensus UUGCGUAUACGCACUGUUAUCAUACUACGUAUACGCACUGUUAUCACAUUACGUAUACGCACUGUAUUCUAAUUACGAA_AGUUG__UUUU_GCUC_UUUUA_ ..(((((((((................)))))))))................................................................... (-11.01 = -11.05 + 0.04)

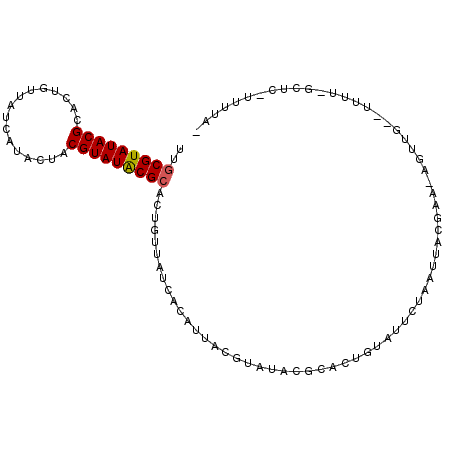

| Location | 10,088,272 – 10,088,374 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 61.22 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088272 102 - 22224390 -UAAAAUGAGCCAAAAGUCAACUUUUCGUAAUUAGAAUACAGGGCGUAUACGUAAUGCCAUAACAGUGCGUAUACGUAGUGUGAAAAUAGUGCGUAUACGCAA -........((.(((((....))))).)).............(((((.......))))).......((((((((((((.(((....))).)))))))))))). ( -31.30) >DroSec_CAF1 4635 102 - 1 -UAAAAAGAGCGAAAAAGCAACUCUUCGUAAUCAGAAUACAGGGCGUAUACGUAAUAUUGUAACAGUGCGUAUACGUAUUAUAAUUACAUUGCGUAUACGCAA -........(((((..........)))))..............(((((((((((((((((((...(((((....)))))))))))...))))))))))))).. ( -27.10) >DroSim_CAF1 4080 78 - 1 -UAAAAAGAGCCAAAAAGCAACUCUUCGUAAUCAG------------------AAU------ACAGGGCGUAUACGUAACAUUGUAACAGUCCGUAUACGCAA -((..(((((...........)))))..)).....------------------...------.....(((((((((....((((...)))).))))))))).. ( -14.90) >DroEre_CAF1 10645 81 - 1 -UAAAA---------------------GUAAUAUGAACACAGUGCGUAUACGCAAUGUGACAACAGUGCGUAUACGUGAUAGGAUAACAGCGCGUAUACGCAA -.....---------------------..........((((.((((....)))).)))).......((((((((((((...(.....)..)))))))))))). ( -28.90) >DroYak_CAF1 3391 82 - 1 AUUUAC---------------------GUAGUUUGAUAACAGUGCGUAUACGUCGUAUGAUAACAGUGCGCAUACGUAGUUUGAUAACAGUGCGUAUACGUCG ......---------------------...(((....)))...((((((((((((((((...........))))))..(((....)))...)))))))))).. ( -17.80) >consensus _UAAAA_GAGC_AAAA__CAACU_UUCGUAAUCAGAAUACAGGGCGUAUACGUAAUAUGAUAACAGUGCGUAUACGUAAUAUGAUAACAGUGCGUAUACGCAA ...........................................(((....)))..............(((((((((((............))))))))))).. (-14.05 = -14.00 + -0.05)

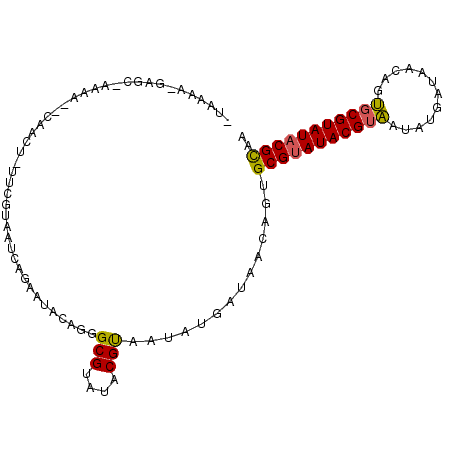

| Location | 10,088,298 – 10,088,399 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.46 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -10.04 |
| Energy contribution | -9.60 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088298 101 + 22224390 ACGUAUACGCACUGUUAUGGCAUUACGUAUACGCCCUGUAUUCUAAUUACGAAAAGUUGACUUUUGGCUCAUUUUAACCGUA-AUUUCACUGUAUAUAAUUA---------- .((((((((...(((....)))...))))))))...(((((...((((((.(((((....)))))((..........)))))-))).....)))))......---------- ( -17.80) >DroSec_CAF1 4661 111 + 1 ACGUAUACGCACUGUUACAAUAUUACGUAUACGCCCUGUAUUCUGAUUACGAAGAGUUGCUUUUUCGCUCUUUUUAACUGUA-AUUUUACUGUGUAUAAUUAUUUGUAUACA .((((((((...(((....)))...))))))))....(((....((((((((((((..((......)).))))))....)))-))).))).((((((((....)))))))). ( -23.10) >DroEre_CAF1 10671 81 + 1 ACGUAUACGCACUGUUGUCACAUUGCGUAUACGCACUGUGUUCAUAUUAC---------------------UUUUAACUGUGCAUUUCAUUCUGUUCAAUUG---------- ..(((((((((.(((....))).)))))))))((((.((...........---------------------.....)).))))...................---------- ( -20.99) >consensus ACGUAUACGCACUGUUAUAACAUUACGUAUACGCCCUGUAUUCUAAUUACGAA_AGUUG__UUUU_GCUC_UUUUAACUGUA_AUUUCACUGUGUAUAAUUA__________ .((((((((...(((....)))...))))))))............................................................................... (-10.04 = -9.60 + -0.44)

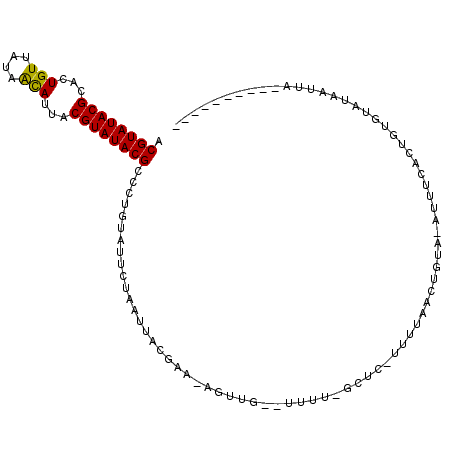

| Location | 10,088,298 – 10,088,399 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.46 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 6.10 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10088298 101 - 22224390 ----------UAAUUAUAUACAGUGAAAU-UACGGUUAAAAUGAGCCAAAAGUCAACUUUUCGUAAUUAGAAUACAGGGCGUAUACGUAAUGCCAUAACAGUGCGUAUACGU ----------................(((-(((((((......))))(((((....))))).))))))..........(((((((((((.((......)).))))))))))) ( -23.80) >DroSec_CAF1 4661 111 - 1 UGUAUACAAAUAAUUAUACACAGUAAAAU-UACAGUUAAAAAGAGCGAAAAAGCAACUCUUCGUAAUCAGAAUACAGGGCGUAUACGUAAUAUUGUAACAGUGCGUAUACGU (((((.(...(((((.(((...))).)))-))..((((..(((((...........)))))..))))..).)))))..(((((((((((............))))))))))) ( -20.60) >DroEre_CAF1 10671 81 - 1 ----------CAAUUGAACAGAAUGAAAUGCACAGUUAAAA---------------------GUAAUAUGAACACAGUGCGUAUACGCAAUGUGACAACAGUGCGUAUACGU ----------...................((((.(((...(---------------------....)...)))...))))(((((((((.(((....))).))))))))).. ( -20.90) >consensus __________UAAUUAUACACAGUGAAAU_UACAGUUAAAA_GAGC_AAAA__CAACU_UUCGUAAUAAGAAUACAGGGCGUAUACGUAAUGUCAUAACAGUGCGUAUACGU ..............................................................................(((((((((((.((......)).))))))))))) (-14.59 = -14.70 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:02 2006