
| Sequence ID | X_DroMel_CAF1 |
|---|---|
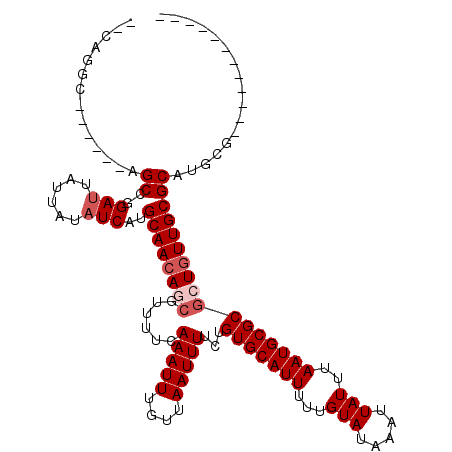
| Location | 10,078,681 – 10,078,781 |
| Length | 100 |
| Max. P | 0.928681 |

| Location | 10,078,681 – 10,078,781 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10078681 100 + 22224390 --CAGGC------AGCCGGAUUAUUAUAUCAUGCAACAGCGUUUUCAAAUUUGUUAAUUUUCUGUGCAUUUUUGUAUAAAUUAUUUAAUGCGCGCUAUUGCGCAUGCG------------ --...((------(((..(((......)))..((((.(((......(((((....)))))...(((((((...(((.....)))..)))))))))).)))))).))).------------ ( -23.20) >DroSec_CAF1 4935 100 + 1 --CAGGC------AGCCGGAUUAUUAUAUCAUGCAACAGCGUUUUCAAAUUUGUUAAUUUUCUGUGCAUUUUUGUAUAAAUUAUUUAAUGCGCGCUGUUGCGCAUGCG------------ --...((------(((..(((......)))..((((((((......(((((....)))))...(((((((...(((.....)))..))))))))))))))))).))).------------ ( -29.20) >DroSim_CAF1 4685 100 + 1 --CAGGC------AGCCGGAUUAUUAUAUCAUGCAACAGCGUUUUCAAAUUUGUUAAUUUUCUGUGCAUUUUUGUAUAAAUUAUUUAAUGCGCGCUGUUGCGCAUGCG------------ --...((------(((..(((......)))..((((((((......(((((....)))))...(((((((...(((.....)))..))))))))))))))))).))).------------ ( -29.20) >DroEre_CAF1 4987 106 + 1 --CAGGCAGCGGCAGCCGGACUAUUAUAUCAUGCAACAUCGUUUUCAAAUUUGUUAAUUUUCUGUGCAUUUUUGUAUAAAUUAUUUAAUGCGCGCUGUUGCGCAUGCG------------ --...((.((((((((.(((..((((...((....................)).))))..)))(((((((...(((.....)))..))))))))))))))))).....------------ ( -25.25) >DroPer_CAF1 4548 113 + 1 AGUCGUC------AGCCGGAUUAUUAUAUCAUGCAACAUAA-UUUGAAAUUUGUUAAUUUUGUGUGCAUUUUUGUAUAAAUUAUUUAAUGCGCGUUGUUGCGCAUGUUACCCCGACCCCA .((((..------..........((((((.(((((.(((((-.((((......))))..))))))))))....))))))........(((((((....))))))).......)))).... ( -23.90) >consensus __CAGGC______AGCCGGAUUAUUAUAUCAUGCAACAGCGUUUUCAAAUUUGUUAAUUUUCUGUGCAUUUUUGUAUAAAUUAUUUAAUGCGCGCUGUUGCGCAUGCG____________ ..............((..(((......)))..((((((((......(((((....)))))...(((((((...(((.....)))..)))))))))))))))))................. (-20.68 = -21.68 + 1.00)


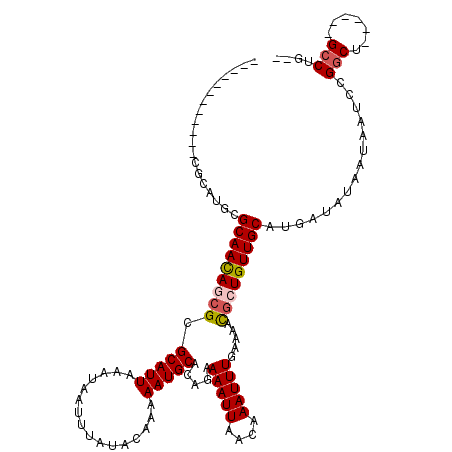
| Location | 10,078,681 – 10,078,781 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -17.43 |
| Energy contribution | -17.91 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10078681 100 - 22224390 ------------CGCAUGCGCAAUAGCGCGCAUUAAAUAAUUUAUACAAAAAUGCACAGAAAAUUAACAAAUUUGAAAACGCUGUUGCAUGAUAUAAUAAUCCGGCU------GCCUG-- ------------.(((.(((((((((((.(((((................))))).((((...........))))....)))))))))..(((......)))..)))------))...-- ( -22.09) >DroSec_CAF1 4935 100 - 1 ------------CGCAUGCGCAACAGCGCGCAUUAAAUAAUUUAUACAAAAAUGCACAGAAAAUUAACAAAUUUGAAAACGCUGUUGCAUGAUAUAAUAAUCCGGCU------GCCUG-- ------------.(((.(((((((((((.(((((................))))).((((...........))))....)))))))))..(((......)))..)))------))...-- ( -23.99) >DroSim_CAF1 4685 100 - 1 ------------CGCAUGCGCAACAGCGCGCAUUAAAUAAUUUAUACAAAAAUGCACAGAAAAUUAACAAAUUUGAAAACGCUGUUGCAUGAUAUAAUAAUCCGGCU------GCCUG-- ------------.(((.(((((((((((.(((((................))))).((((...........))))....)))))))))..(((......)))..)))------))...-- ( -23.99) >DroEre_CAF1 4987 106 - 1 ------------CGCAUGCGCAACAGCGCGCAUUAAAUAAUUUAUACAAAAAUGCACAGAAAAUUAACAAAUUUGAAAACGAUGUUGCAUGAUAUAAUAGUCCGGCUGCCGCUGCCUG-- ------------.(((.(((...((((.((.((((......(((((.....(((((((.....((((.....))))......)).)))))..))))))))).)))))).))))))...-- ( -18.50) >DroPer_CAF1 4548 113 - 1 UGGGGUCGGGGUAACAUGCGCAACAACGCGCAUUAAAUAAUUUAUACAAAAAUGCACACAAAAUUAACAAAUUUCAAA-UUAUGUUGCAUGAUAUAAUAAUCCGGCU------GACGACU ...(((((((.....((((((......))))))........(((((.....(((((.(((.((((...........))-)).))))))))..)))))...)))))))------....... ( -24.40) >consensus ____________CGCAUGCGCAACAGCGCGCAUUAAAUAAUUUAUACAAAAAUGCACAGAAAAUUAACAAAUUUGAAAACGCUGUUGCAUGAUAUAAUAAUCCGGCU______GCCUG__ ...................(((((((((.(((((................))))).....(((((....))))).....)))))))))...............(((.......))).... (-17.43 = -17.91 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:52 2006