
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,064,734 – 10,064,870 |
| Length | 136 |
| Max. P | 0.919468 |

| Location | 10,064,734 – 10,064,841 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.00 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10064734 107 - 22224390 CUUCACUGAAAUUUGCAUAAAUUUGCAUUAUUUUCGGUGAACCGAAAAUCAUAAUGGGCGUGGACAAUGGGCUCAUAAAUUGAGUUUAGCCAA-AAAUAGAAAAAGCC .((((((((((..((((......))))....))))))))))...............(((.(((....((((((((.....)))))))).))).-...........))) ( -28.32) >DroSec_CAF1 1278 107 - 1 CAUCACUGAAAUUUGCAUAAAUUUGCAAAAUUUUCGGUGAACCGAAAAUCAUAAUGGGCGUGGACAAUGGGCUCAUAAAUUGAGUUUAGCCGA-AAAUAGAAGAGGCC ..((((.....((((((......))))))((((((((....))))))))..........))))....((((((((.....))))))))(((..-..........))). ( -29.00) >DroEre_CAF1 865 104 - 1 CAUUACGGAAAUUUGCAUAAAUUUGCAUAAUUUUCGGUGAACCAAAAAUCAUAAUGGGUGUGGACAAUGGGCUCAUAAA----GCUUGCCCAAAAAAUGAAAGAAACC ((((.((((((((((((......)))).)))))))).....(((...(((......))).)))....(((((.......----....)))))...))))......... ( -24.00) >consensus CAUCACUGAAAUUUGCAUAAAUUUGCAUAAUUUUCGGUGAACCGAAAAUCAUAAUGGGCGUGGACAAUGGGCUCAUAAAUUGAGUUUAGCCAA_AAAUAGAAGAAGCC ..(((((((((..((((......))))....)))))))))................(((.(((....((((((((.....)))))))).))).............))) (-21.53 = -21.54 + 0.01)

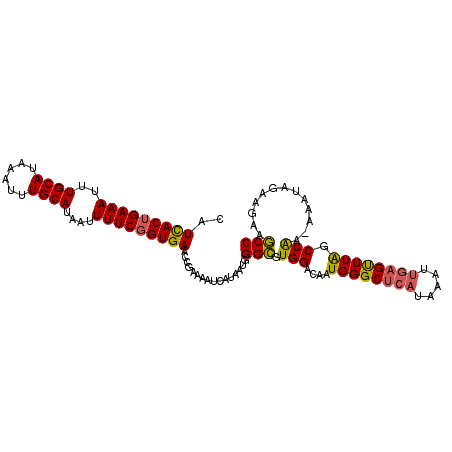

| Location | 10,064,770 – 10,064,870 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -15.89 |
| Consensus MFE | -10.93 |
| Energy contribution | -12.30 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10064770 100 + 22224390 CCCAUUGUCCACGCCCAUUAUGAUUUUCGGUUCACCGAAAAUAAUGCAAAUUUAUGCAAAUUUCAGUGAAGCUUC---CUCAACACAAAUAGC--------GCAAAAACAC ....((((.(((..........((((((((....))))))))..((((......)))).......)))..(((..---............)))--------))))...... ( -17.64) >DroSec_CAF1 1314 103 + 1 CCCAUUGUCCACGCCCAUUAUGAUUUUCGGUUCACCGAAAAUUUUGCAAAUUUAUGCAAAUUUCAGUGAUGCUUCCUCCUCAACACAAAUAGC--------GCAAAAACAC ....((((.(((.........(((((((((....)))))))))(((((......)))))......)))..(((.................)))--------))))...... ( -18.53) >DroEre_CAF1 898 108 + 1 CCCAUUGUCCACACCCAUUAUGAUUUUUGGUUCACCGAAAAUUAUGCAAAUUUAUGCAAAUUUCCGUAAUGCUUC---CUCAACCCAAACAGCACAAAAACGCAAAAACAC ...............(((((((((((((((....))))))))..((((......))))......)))))))....---.............((........))........ ( -16.10) >DroYak_CAF1 32410 98 + 1 CCCAUUGUCCACACCCAUUAUGAUAUUCGGUUCACCAAAAAUGAUGCAAAUUUAUGCAAAUUCCCGUGAGGCUUC---CUCAAACGAAA----------AAAAAAAAACAC .........................((((...............((((......))))........(((((...)---))))..)))).----------............ ( -11.30) >consensus CCCAUUGUCCACACCCAUUAUGAUUUUCGGUUCACCGAAAAUUAUGCAAAUUUAUGCAAAUUUCAGUGAUGCUUC___CUCAACACAAAUAGC________GCAAAAACAC .........(((.......(((((((((((....)))))))))))(((......)))........)))........................................... (-10.93 = -12.30 + 1.37)

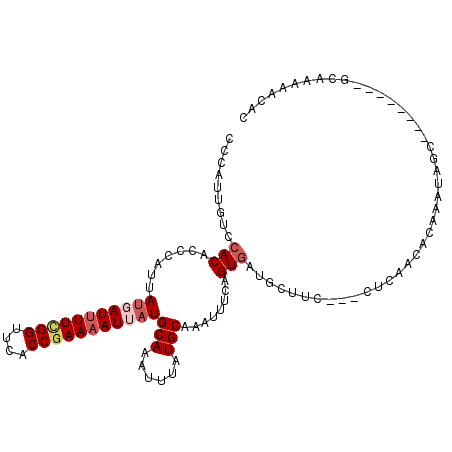

| Location | 10,064,770 – 10,064,870 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10064770 100 - 22224390 GUGUUUUUGC--------GCUAUUUGUGUUGAG---GAAGCUUCACUGAAAUUUGCAUAAAUUUGCAUUAUUUUCGGUGAACCGAAAAUCAUAAUGGGCGUGGACAAUGGG .....((..(--------(((..(((((((..(---(....((((((((((..((((......))))....))))))))))))...)).)))))..))))..))....... ( -29.60) >DroSec_CAF1 1314 103 - 1 GUGUUUUUGC--------GCUAUUUGUGUUGAGGAGGAAGCAUCACUGAAAUUUGCAUAAAUUUGCAAAAUUUUCGGUGAACCGAAAAUCAUAAUGGGCGUGGACAAUGGG .....((..(--------(((..(((((((..((.(....).(((((((((((((((......))))))..))))))))).))...)).)))))..))))..))....... ( -30.60) >DroEre_CAF1 898 108 - 1 GUGUUUUUGCGUUUUUGUGCUGUUUGGGUUGAG---GAAGCAUUACGGAAAUUUGCAUAAAUUUGCAUAAUUUUCGGUGAACCAAAAAUCAUAAUGGGUGUGGACAAUGGG .((((..(((.(..(((((...((((((((...---..)))....((((((((((((......)))).)))))))).....)))))...))))).).)))..))))..... ( -25.90) >DroYak_CAF1 32410 98 - 1 GUGUUUUUUUUU----------UUUCGUUUGAG---GAAGCCUCACGGGAAUUUGCAUAAAUUUGCAUCAUUUUUGGUGAACCGAAUAUCAUAAUGGGUGUGGACAAUGGG ((((.......(----------(..(((..(((---(...)))))))..))...))))...((..((((...(((((....)))))..........))))..))....... ( -20.02) >consensus GUGUUUUUGC________GCUAUUUGGGUUGAG___GAAGCAUCACGGAAAUUUGCAUAAAUUUGCAUAAUUUUCGGUGAACCGAAAAUCAUAAUGGGCGUGGACAAUGGG .((((..(((................................(((((((((..((((......))))....))))))))).(((..........))))))..))))..... (-16.96 = -16.65 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:48 2006