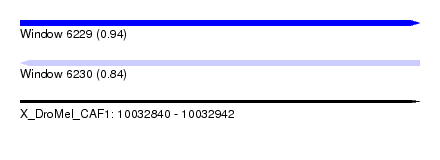
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,032,840 – 10,032,942 |
| Length | 102 |
| Max. P | 0.937847 |

| Location | 10,032,840 – 10,032,942 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -16.83 |
| Energy contribution | -18.17 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10032840 102 + 22224390 AUGAAAACUGACAACAAGGACCUUUAUU-UCACAACGUAGGCCGUUGGAAAUGG--GAAAACAACGUUUAGCACUUUAAAGUGCAGUCAGUAUUGUACUAGCAUU ......((((((........(((..(((-((.(((((.....))))))))))))--).............(((((....))))).)))))).............. ( -26.20) >DroSec_CAF1 1024 101 + 1 AUGAAGACUGACAACCAGGACCUUUAUU-CCACAACGUAGGCCGUUGGAAGUGGAAGAAAACAAGGUUUAGCACUUUAAAGUGCAGUCAGUAUCGUACCAG---U ((((..((((((.....(((((((..((-((((((((.....))))....))))))......))))))).(((((....))))).)))))).)))).....---. ( -30.90) >DroYak_CAF1 1080 92 + 1 AUAAAGACUGUCAGCAAGGACCUUUGCUGCCACAACGUAGGCCGUAGAAAAUGG--UAAAACAUGGUUUGGCACUUUAAAAUGCAGUCAGUAU-----------U .....((((((.((((((....))))))((((...(((..(((((.....))))--).....)))...))))..........)))))).....-----------. ( -24.70) >consensus AUGAAGACUGACAACAAGGACCUUUAUU_CCACAACGUAGGCCGUUGGAAAUGG__GAAAACAAGGUUUAGCACUUUAAAGUGCAGUCAGUAU_GUAC_AG___U ......((((((.....(((((..........(((((.....)))))....((........)).))))).(((((....))))).)))))).............. (-16.83 = -18.17 + 1.33)

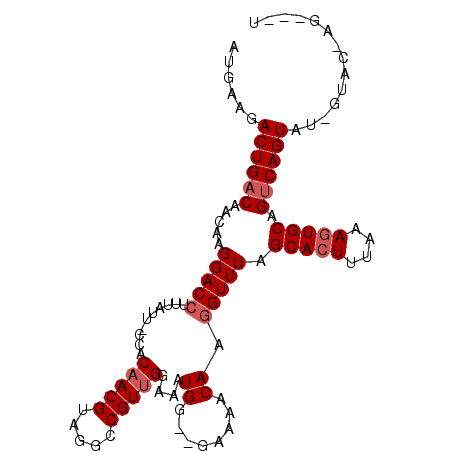
| Location | 10,032,840 – 10,032,942 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -16.93 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10032840 102 - 22224390 AAUGCUAGUACAAUACUGACUGCACUUUAAAGUGCUAAACGUUGUUUUC--CCAUUUCCAACGGCCUACGUUGUGA-AAUAAAGGUCCUUGUUGUCAGUUUUCAU ...(((.(.((((((..(((((((((....)))))..............--..((((((((((.....))))).))-)))...))))..))))))))))...... ( -25.10) >DroSec_CAF1 1024 101 - 1 A---CUGGUACGAUACUGACUGCACUUUAAAGUGCUAAACCUUGUUUUCUUCCACUUCCAACGGCCUACGUUGUGG-AAUAAAGGUCCUGGUUGUCAGUCUUCAU .---.......((.((((((.(((((....)))))..((((...((((.((((((....((((.....))))))))-)).)))).....))))))))))..)).. ( -28.30) >DroYak_CAF1 1080 92 - 1 A-----------AUACUGACUGCAUUUUAAAGUGCCAAACCAUGUUUUA--CCAUUUUCUACGGCCUACGUUGUGGCAGCAAAGGUCCUUGCUGACAGUCUUUAU .-----------(((..(((((......((((((..((((...))))..--.))))))(((((((....)))))))((((((......)))))).)))))..))) ( -19.50) >consensus A___CU_GUAC_AUACUGACUGCACUUUAAAGUGCUAAACCUUGUUUUC__CCAUUUCCAACGGCCUACGUUGUGG_AAUAAAGGUCCUUGUUGUCAGUCUUCAU ..............((((((.(((((....)))))...........................(((((...((((....)))))))))......))))))...... (-16.93 = -16.60 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:41 2006