
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,030,688 – 10,030,799 |
| Length | 111 |
| Max. P | 0.867019 |

| Location | 10,030,688 – 10,030,799 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.11 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
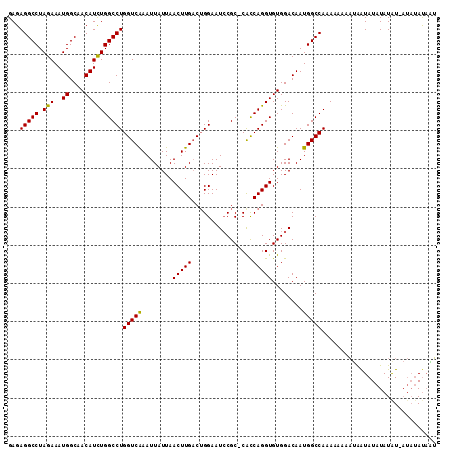
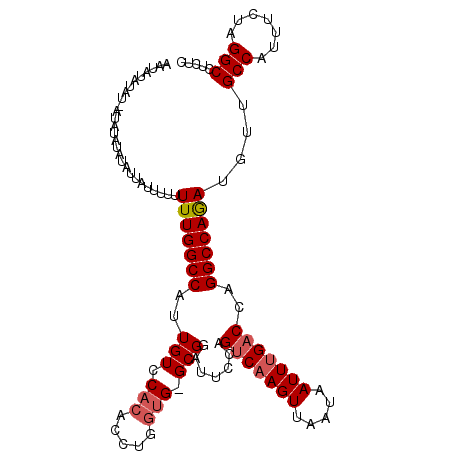
>X_DroMel_CAF1 10030688 111 + 22224390 GAGAGGCCUAGAAAUGGCAACAUUUGGCCUGGUCAAAUUAUUAACUUGCCGGGAAUCCGCCCAUCAGGUGUGGACAAUGGCCAAAAAAAAUGAUAUAUAUAUUAUAUAC-AU ....((((...(((((....)))))..(((((.(((.........))))))))..(((((((....)).)))))....)))).......((((((((.....))))).)-)) ( -31.60) >DroSim_CAF1 6869 108 + 1 GAGAGGCCUAGAAAUGGCAACAUCUGGCCUGGUCAAAUUAUUAACUUGACUGGAAUGCGC-CACCAGGUGUGGACAAUGGCCAAAAA--AUAAUAUAUAUGU-AUAUAUAGU ...(((((.(((..((....))))))))))(((((........(((((..(((......)-)).)))))........))))).....--....((((((...-.)))))).. ( -27.59) >DroEre_CAF1 37984 106 + 1 GAGAGGCCUAGAAAUGGCCACAUCUGGCCUGGUCGAAUUAUUAACUUGACUGGAAUCCGC-CGCCAGGUGUGGACAAUGGCCAAAAAAAUAAGUAUCUAUAU-----AUAUG ....((((......((.(((((((((((..((((((.........))))))((......)-)))))))))))).))..))))....................-----..... ( -34.10) >consensus GAGAGGCCUAGAAAUGGCAACAUCUGGCCUGGUCAAAUUAUUAACUUGACUGGAAUCCGC_CACCAGGUGUGGACAAUGGCCAAAAAAAAUAAUAUAUAUAU_AUAUAUAAU ...(((((.(((..((....))))))))))(((((........(((((................)))))........))))).............................. (-20.55 = -20.11 + -0.44)


| Location | 10,030,688 – 10,030,799 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -17.02 |
| Energy contribution | -18.13 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
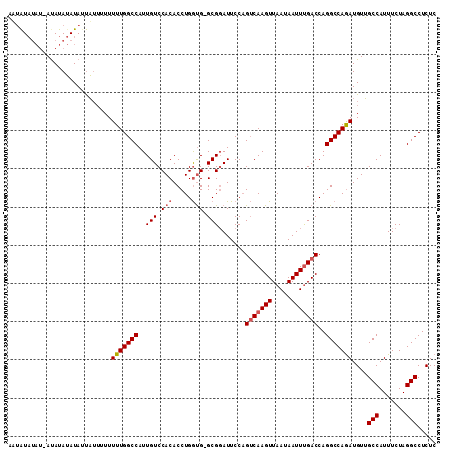
>X_DroMel_CAF1 10030688 111 - 22224390 AU-GUAUAUAAUAUAUAUAUCAUUUUUUUUGGCCAUUGUCCACACCUGAUGGGCGGAUUCCCGGCAAGUUAAUAAUUUGACCAGGCCAAAUGUUGCCAUUUCUAGGCCUCUC ((-((((((...)))))))).......(((((((...((((((....).)))))((....))(((((((.....))))).)).))))))).(..(((.......)))..).. ( -27.60) >DroSim_CAF1 6869 108 - 1 ACUAUAUAU-ACAUAUAUAUUAU--UUUUUGGCCAUUGUCCACACCUGGUG-GCGCAUUCCAGUCAAGUUAAUAAUUUGACCAGGCCAGAUGUUGCCAUUUCUAGGCCUCUC ..((((((.-...))))))....--..(((((((..(((((((.....)))-).))).....(((((((.....)))))))..))))))).(..(((.......)))..).. ( -27.20) >DroEre_CAF1 37984 106 - 1 CAUAU-----AUAUAGAUACUUAUUUUUUUGGCCAUUGUCCACACCUGGCG-GCGGAUUCCAGUCAAGUUAAUAAUUCGACCAGGCCAGAUGUGGCCAUUUCUAGGCCUCUC .....-----....................((((..((.(((((.((((((-(((((((..............)))))).))..))))).))))).))......)))).... ( -28.54) >consensus AAUAUAUAU_AUAUAUAUAUUAUUUUUUUUGGCCAUUGUCCACACCUGGUG_GCGGAUUCCAGUCAAGUUAAUAAUUUGACCAGGCCAGAUGUUGCCAUUUCUAGGCCUCUC ...........................(((((((..(((.(((.....))).))).......(((((((.....)))))))..)))))))....(((.......)))..... (-17.02 = -18.13 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:38 2006