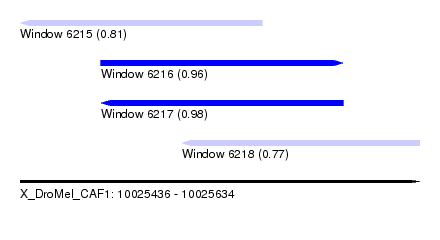
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,025,436 – 10,025,634 |
| Length | 198 |
| Max. P | 0.983994 |

| Location | 10,025,436 – 10,025,556 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10025436 120 - 22224390 AUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCUAGAUGCCCAAGUGGGCGGUGCCAGAAGAGGAAACAAGUAAAGUGUAUUAGAGAUACUCUGCACAGCAAGAGGCAUAUUUGG ..................((((....................(((((....)))))((((.(((...(....).........(((((...))))))))))))......))))........ ( -25.50) >DroSim_CAF1 2763 96 - 1 AUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAA------------------------GCCAGCAGAGGAAACAACCAAAGUGUAUUAGAGAUACUCUGUACAGCAAGAGGCAUGUUUAA ....(((((((.(((((........)))))....------------------------(((.((...(....)........(((((.((((...)))))))))))....))).))))))) ( -14.30) >DroEre_CAF1 32864 114 - 1 AUAAUUCAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCCACAUGCCCAACUGGGCGG------AAGAGGAAACAACUAAAGUGUAUUAGAGAUACUCUGUGCAGCAAGAGGCAUGUUUAA .......................................((((((((..(((.((((------(...(....).........(((((...)))))))))).)))...).))))))).... ( -23.30) >DroYak_CAF1 30140 108 - 1 AUAAUUAAAUAAAUUAAAGCCUAAGUUAAUCAGAAAGCCACAUGCUCAAUUGGGCGA------U------GUCAACUAAAGUGUAUUAGAGAUACUCUGUACAGCAAAAGGCAUGUUGAA .............................((((...(((((((((((....)))).)------)------)).........(((((.((((...)))))))))......)))...)))). ( -20.50) >consensus AUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCCACAUGCCCAA_UGGGCGG______AAGAGGAAACAACUAAAGUGUAUUAGAGAUACUCUGUACAGCAAGAGGCAUGUUUAA ..................((((....................(((((....))))).........................(((((.((((...))))))))).....))))........ (-15.23 = -15.47 + 0.25)


| Location | 10,025,476 – 10,025,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10025476 120 + 22224390 UUUACUUGUUUCCUCUUCUGGCACCGCCCACUUGGGCAUCUAGCUUUUUGAUUAAUUUAGGCUUUAAUUUAUUUAAUUAUAAGCCAAAGGCAUAGGGAAUUUUGUCACCUGUUGGCACCC .......(((((((...........((((....)))).....(((((((((.....)))((((((((((.....))))).)))))))))))..)))))))..(((((.....)))))... ( -28.20) >DroSim_CAF1 2803 96 + 1 UUUGGUUGUUUCCUCUGCUGGC------------------------UUUGAUUAAUUUAGGCUUUAAUUUAUUUAAUUAUAAGCCAAAGGCACAGGGAAUUUUGUCACCUGUUGGCACCC ...(((.(((((((.(((((((------------------------(((((((((..((((......)))).))))))).)))))...)))).)))))))...((((.....))))))). ( -24.20) >DroEre_CAF1 32904 114 + 1 UUUAGUUGUUUCCUCUU------CCGCCCAGUUGGGCAUGUGGCUUUUUGAUUAAUUUAGGCUUUAAUUUAUUGAAUUAUAAGCCACAGGCAUAGGGAAUUUUGUCACCUGUUGGCAUCC .......(((((((...------((((((....)))).((((((((......((((((((...........)))))))).))))))))))...)))))))..(((((.....)))))... ( -32.90) >DroYak_CAF1 30180 108 + 1 UUUAGUUGAC------A------UCGCCCAAUUGAGCAUGUGGCUUUCUGAUUAACUUAGGCUUUAAUUUAUUUAAUUAUAAGCCCAAGGCAUAGGGAAUUUUAUCACCUGUUGGCAGCC ....((..((------.------..(((((...((((.....))))..)).........((((((((((.....))))).)))))...)))..(((((......)).)))))..)).... ( -24.60) >consensus UUUAGUUGUUUCCUCUU______CCGCCCA_UUGGGCAUGUGGCUUUUUGAUUAAUUUAGGCUUUAAUUUAUUUAAUUAUAAGCCAAAGGCAUAGGGAAUUUUGUCACCUGUUGGCACCC .........................((((....))))......................((((((((((.....))))).)))))...(.((((((((......)).)))).)).).... (-17.84 = -18.15 + 0.31)

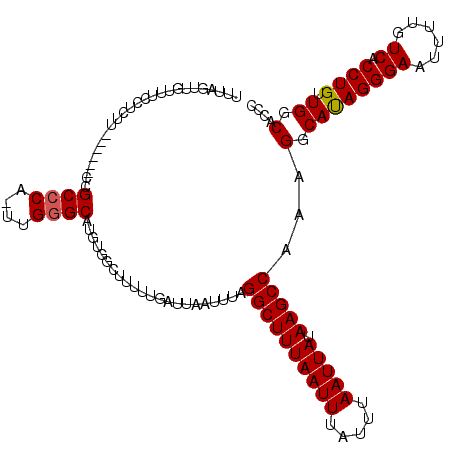
| Location | 10,025,476 – 10,025,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10025476 120 - 22224390 GGGUGCCAACAGGUGACAAAAUUCCCUAUGCCUUUGGCUUAUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCUAGAUGCCCAAGUGGGCGGUGCCAGAAGAGGAAACAAGUAAA .((..((...(((.((......)))))........(((((.(((((.....))))))))))......................((((....))))))..))......(....)....... ( -29.50) >DroSim_CAF1 2803 96 - 1 GGGUGCCAACAGGUGACAAAAUUCCCUGUGCCUUUGGCUUAUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAA------------------------GCCAGCAGAGGAAACAACCAAA .((((((....))).......((((((((..(((((((((.(((((.....))))))))))..........)))------------------------)...)))).))))...)))... ( -21.10) >DroEre_CAF1 32904 114 - 1 GGAUGCCAACAGGUGACAAAAUUCCCUAUGCCUGUGGCUUAUAAUUCAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCCACAUGCCCAACUGGGCGG------AAGAGGAAACAACUAAA ....(((....))).......((((((...((((((((((..................................)))))))).((((....))))))------.)).))))......... ( -25.68) >DroYak_CAF1 30180 108 - 1 GGCUGCCAACAGGUGAUAAAAUUCCCUAUGCCUUGGGCUUAUAAUUAAAUAAAUUAAAGCCUAAGUUAAUCAGAAAGCCACAUGCUCAAUUGGGCGA------U------GUCAACUAAA ((((..(...(((.((......)))))....(((((((((.(((((.....)))))))))))))).......)..))))((((((((....)))).)------)------))........ ( -30.20) >consensus GGGUGCCAACAGGUGACAAAAUUCCCUAUGCCUUUGGCUUAUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCCACAUGCCCAA_UGGGCGG______AAGAGGAAACAACUAAA .(..(((....)))..)....((((..........(((((.(((((.....)))))))))).....................(((((....)))))...........))))......... (-14.14 = -14.70 + 0.56)

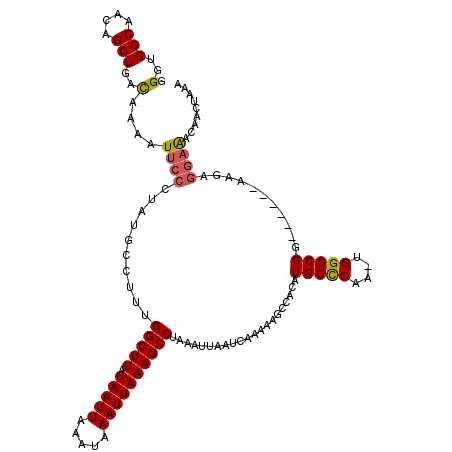
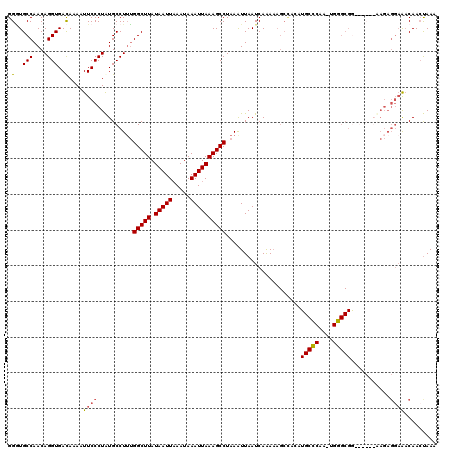
| Location | 10,025,516 – 10,025,634 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -12.54 |
| Energy contribution | -12.85 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10025516 118 - 22224390 AAAACCUUUCUCGCACAC--ACACACACACACGAAUUUUUGGGUGCCAACAGGUGACAAAAUUCCCUAUGCCUUUGGCUUAUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCUA ..................--............((...((((((((((((.((((...............)))))))))...(((((.....)))))..)))))))....))......... ( -16.96) >DroSim_CAF1 2825 106 - 1 AAAAUCUUUCG------C--ACACACACACACGAAUUUUUGGGUGCCAACAGGUGACAAAAUUCCCUGUGCCUUUGGCUUAUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAA------ ...........------.--............((...((((((((((((.((((.(((........))))))))))))...(((((.....)))))..)))))))....))...------ ( -19.60) >DroEre_CAF1 32938 110 - 1 AAAACCCUUCGAGCACAC----------ACACAAUUUUUUGGAUGCCAACAGGUGACAAAAUUCCCUAUGCCUGUGGCUUAUAAUUCAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCCA ..................----------......((((((((......((((((...............))))))(((((.(((((.....))))))))))........))))))))... ( -17.36) >DroYak_CAF1 30208 120 - 1 AAAACCUUUCGCCCACACACACACACACACACGAAUUUUUGGCUGCCAACAGGUGAUAAAAUUCCCUAUGCCUUGGGCUUAUAAUUAAAUAAAUUAAAGCCUAAGUUAAUCAGAAAGCCA .....(((((......................(((((((...(.(((....))))..))))))).......(((((((((.(((((.....)))))))))))))).......)))))... ( -23.50) >consensus AAAACCUUUCGCGCACAC__ACACACACACACGAAUUUUUGGGUGCCAACAGGUGACAAAAUUCCCUAUGCCUUUGGCUUAUAAUUAAAUAAAUUAAAGCCUAAAUUAAUCAAAAAGCCA ...................................(((((((........(((.((......)))))........(((((.(((((.....))))))))))........))))))).... (-12.54 = -12.85 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:27 2006