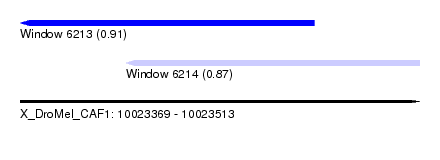
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,023,369 – 10,023,513 |
| Length | 144 |
| Max. P | 0.911599 |

| Location | 10,023,369 – 10,023,475 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.07 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
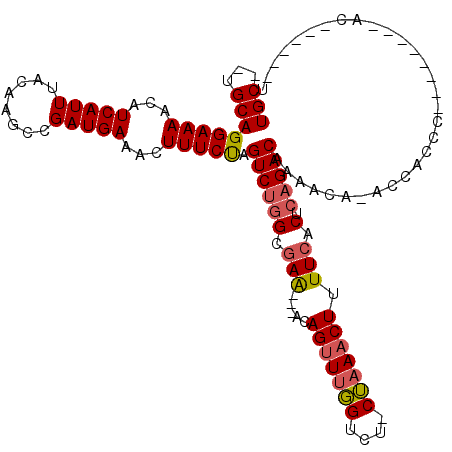
>X_DroMel_CAF1 10023369 106 - 22224390 UCUAGUCUGGCGAAACAGUUUGGUCUCUAAACUUUUCACUCAGACAAAACUAAACACCCACAACCCAUCCUCCAUCCGAAAAUGCUAUCCCUUGCCGGCUUA-CAGC ....((((((.((((.(((((((...))))))))))).).)))))..............................(((.....((........)))))....-.... ( -17.40) >DroEre_CAF1 30594 94 - 1 UCUAGUCUGGCGAAACAGUUUGGUCUCUAAACUUUUCACUCAGACAAAACU-CCAA---AC------UC--CCCCCACAAAAUGCUAUCCUUUGCCGGCAUA-CAGC ....((((((.((((.(((((((...))))))))))).).)))))......-....---..------..--..........(((((..........))))).-.... ( -18.40) >DroYak_CAF1 27620 100 - 1 UCUAGUCUGGCGAAACAGUUUGGUCUCUAAACUUUUCACUCAGACAAAGCA-ACCACCCGC------UCCGCCCCCCUAAAAUGCUAUCCCUUGCCGGCUUGCCAGC ....((((((.((((.(((((((...))))))))))).).)))))...(((-(((....((------...))...........((........)).)).)))).... ( -21.30) >consensus UCUAGUCUGGCGAAACAGUUUGGUCUCUAAACUUUUCACUCAGACAAAACU_ACCACCCAC______UCC_CCCCCCCAAAAUGCUAUCCCUUGCCGGCUUA_CAGC ....((((((.((((.(((((((...))))))))))).).)))))......................................(((..........)))........ (-16.07 = -16.07 + -0.00)



| Location | 10,023,407 – 10,023,513 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.19 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10023407 106 - 22224390 UGC--AGCAGGAAAACAUCAUUUACAAGCCGAUGAAACUUUCUAGUCUGGCGAA---ACAGUUUGGUCU-CUAAACUUUUCACUCAGACAAAACUAAACACCC--------ACAACCCAU ...--....(((((...(((((........)))))...))))).((((((.(((---(.(((((((...-))))))))))).).)))))..............--------......... ( -20.20) >DroAna_CAF1 22483 105 - 1 UGCGGGGCAGGAAAACAUCAUUUACAAGCCGAUGAAAGUUUCCA--CUUGCCAGUUUCCAGCUCCGUCUGCGAAACUUUUCACUUUG------CA-AUCAGCCAGUUGGGGGC------U .....(((((((((...(((((........)))))...))))).--..))))(((..(((((((.(..((((((.........))))------))-..).)..))))))..))------) ( -30.40) >DroEre_CAF1 30630 96 - 1 UGC--UGCAGGAAAACAUCAUUUACAAACCGAUGAAACUUUCUAGUCUGGCGAA---ACAGUUUGGUCU-CUAAACUUUUCACUCAGACAAAACU-CCAA-----------AC------U ...--....(((...((((...........))))..........((((((.(((---(.(((((((...-))))))))))).).))))).....)-))..-----------..------. ( -20.90) >DroYak_CAF1 27659 99 - 1 UGC--UGCAGGAAAACAUCAUUUACAAGCCGAUGAAACUUUCUAGUCUGGCGAA---ACAGUUUGGUCU-CUAAACUUUUCACUCAGACAAAGCA-ACCACCC--------GC------U (((--(...(((((...(((((........)))))...))))).((((((.(((---(.(((((((...-))))))))))).).)))))..))))-.......--------..------. ( -24.00) >consensus UGC__UGCAGGAAAACAUCAUUUACAAGCCGAUGAAACUUUCUAGUCUGGCGAA___ACAGUUUGGUCU_CUAAACUUUUCACUCAGACAAAACA_ACCACCC________AC______U (((...)))(((((...(((((........)))))...))))).((((((.(((.....(((((((....))))))).))).).)))))............................... (-13.23 = -13.85 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:22 2006