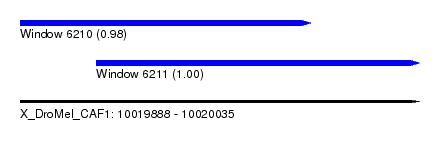
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,019,888 – 10,020,035 |
| Length | 147 |
| Max. P | 0.998372 |

| Location | 10,019,888 – 10,019,995 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.44 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -17.28 |
| Energy contribution | -16.96 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10019888 107 + 22224390 UUACUACUGCUGUUAACCCUCCAAUACCAACGCGACCCCCCCACUACCCCUUUCUUUUUUACAUGUAUAUGUGAAGAGGGGGGGGGGGGGCUUGUG-AGAAAGAAGGU ...............(((.((.........((((((((((((....(((((((((...((((((....)))))))))))))))))))))).)))))-.....)).))) ( -38.64) >DroEre_CAF1 27270 96 + 1 UCACUACUGCUGUUAACCCUCCAAUACCACCACCCCUCCCCC----UCCCUUGCCAUCUUACAAGUAUAUGUGG-----GGGAAGGGGGGC--GUG-CGAAAGGAGCC ..................((((.....((((.((((((((((----.(.((((........)))).....).))-----))).))))).).--)))-.....)))).. ( -30.10) >DroYak_CAF1 23703 95 + 1 UUACUACUGCUGUUAACCCUCCAAUACCAUUCUCUUUUCCCC----UCCCUUACCAUUUUACAUGUAUAAGUGG-----G--AGGGGGGGC--GAGGGGGAAGGAGAC ..............................((((((((((((----(((((..((.((((((........))))-----)--)))..))).--)))))))))))))). ( -36.70) >consensus UUACUACUGCUGUUAACCCUCCAAUACCAACACCACUCCCCC____UCCCUUACCAUUUUACAUGUAUAUGUGG_____GGGAGGGGGGGC__GUG_AGAAAGGAGAC ..................((((........(((...((((((....((((........((((........)))).....)))).))))))...)))......)))).. (-17.28 = -16.96 + -0.32)


| Location | 10,019,916 – 10,020,035 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.41 |
| Mean single sequence MFE | -47.50 |
| Consensus MFE | -24.23 |
| Energy contribution | -23.03 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10019916 119 + 22224390 AACGCGACCCCCCCACUACCCCUUUCUUUUUUACAUGUAUAUGUGAAGAGGGGGGGGGGGGGCUUGUG-AGAAAGAAGGUCGGAAAUGGGGGCGGGGUCGAGGCAAAAACGAUGUGGAGC ..(.((((((((((.((((((((..(((((((((((....)))))))))))..))))).((.(((.(.-....).))).)).....)))))).))))))).).................. ( -51.70) >DroEre_CAF1 27298 108 + 1 ACCACCCCUCCCCC----UCCCUUGCCAUCUUACAAGUAUAUGUGG-----GGGAAGGGGGGC--GUG-CGAAAGGAGCCCACAAUUGGGGGCGGGGUCGAGGCAAAAACGAUGUGGAGC .(((((((.(((((----.(((((.((..((((((......)))))-----))))))))((((--.(.-(....).)))))......))))).)))((((.........))))))))... ( -48.40) >DroYak_CAF1 23731 107 + 1 AUUCUCUUUUCCCC----UCCCUUACCAUUUUACAUGUAUAAGUGG-----G--AGGGGGGGC--GAGGGGGAAGGAGACCGGAAUUGGGGGCGGGGUCGAGGCAGCAACGAUGUGAAGC ..((((((((((((----(((((..((.((((((........))))-----)--)))..))).--)))))))))))))).....((((...((...((....)).))..))))....... ( -42.40) >consensus AACACCACUCCCCC____UCCCUUACCAUUUUACAUGUAUAUGUGG_____GGGAGGGGGGGC__GUG_AGAAAGGAGACCGGAAUUGGGGGCGGGGUCGAGGCAAAAACGAUGUGGAGC .(((((((.(((((....((((((.((...((((........)))).....)).))))))..............((...))......))))).)))((((.........))))))))... (-24.23 = -23.03 + -1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:19 2006