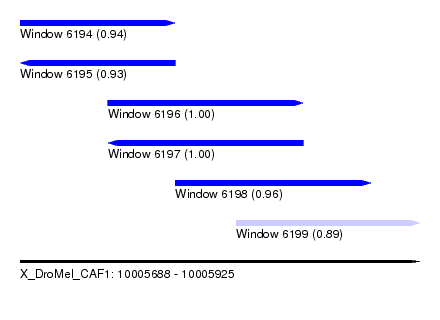
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,005,688 – 10,005,925 |
| Length | 237 |
| Max. P | 0.998747 |

| Location | 10,005,688 – 10,005,780 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10005688 92 + 22224390 UAACUGCAAAAUGUAAUUUUGUGGAAAUCACAAAAUGUGUGAGUGUGUGUGGAAUGAGAGCCGCACGCAAACACACAGAAUGCCCACACUGC .....(((.......((((((((.....))))))))(((((..(((((((((........)))))))))..)))))....)))......... ( -31.20) >DroSec_CAF1 11232 92 + 1 UAACUGCAAAAUGUAAUUUUGUGGAAAUCACAAAAUGUUUGAGUGUGUGUGGGAUGAGGACCACACGCAAACACACAGAAUGCCCACACUGC .....(((.......((((((((.....))))))))((.((..(((((((((........)))))))))..)).))....)))......... ( -26.70) >DroYak_CAF1 8422 87 + 1 UAACUGCAAAAUGUAAUUUUGUGGAAAUCACAAAACGAGUGUGUGUGUGU-----GAGAGCCACACGCAAACAAGCGGAGUACCCACACUGC ...((((.........(((((((.....))))))).(..((((((((.((-----....))))))))))..)..)))).............. ( -26.20) >consensus UAACUGCAAAAUGUAAUUUUGUGGAAAUCACAAAAUGUGUGAGUGUGUGUGG_AUGAGAGCCACACGCAAACACACAGAAUGCCCACACUGC .....(((...(((..(((((((.....)))))))(((.((..(((((((((........)))))))))..)).)))........))).))) (-21.73 = -22.07 + 0.34)



| Location | 10,005,688 – 10,005,780 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10005688 92 - 22224390 GCAGUGUGGGCAUUCUGUGUGUUUGCGUGCGGCUCUCAUUCCACACACACUCACACAUUUUGUGAUUUCCACAAAAUUACAUUUUGCAGUUA ((((.(((........(((((..((.(((.((........)).))).))..)))))((((((((.....))))))))..))).))))..... ( -27.70) >DroSec_CAF1 11232 92 - 1 GCAGUGUGGGCAUUCUGUGUGUUUGCGUGUGGUCCUCAUCCCACACACACUCAAACAUUUUGUGAUUUCCACAAAAUUACAUUUUGCAGUUA ((((.(((........((.((..((.((((((........)))))).))..)).))((((((((.....))))))))..))).))))..... ( -26.10) >DroYak_CAF1 8422 87 - 1 GCAGUGUGGGUACUCCGCUUGUUUGCGUGUGGCUCUC-----ACACACACACACUCGUUUUGUGAUUUCCACAAAAUUACAUUUUGCAGUUA ((((((((((....))((......))(((((......-----.))))).)))))..((((((((.....)))))))).......)))..... ( -26.20) >consensus GCAGUGUGGGCAUUCUGUGUGUUUGCGUGUGGCUCUCAU_CCACACACACUCACACAUUUUGUGAUUUCCACAAAAUUACAUUUUGCAGUUA ((((((((((....(((..((....))..))).......)))))))..........((((((((.....)))))))).......)))..... (-20.70 = -20.93 + 0.23)

| Location | 10,005,740 – 10,005,856 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -44.82 |
| Consensus MFE | -39.29 |
| Energy contribution | -39.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -5.02 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10005740 116 + 22224390 AAUGAGAGCCGCACGCAAACACACAGAAUGCCCACACUGCGUAUGAGUAAUCCGACUGGCAAGCAAUUGCGUUACUCAUACGCAGUGUGGGUCGCCGACGACCAACCCCCCGUAAA ..........(((..(.........)..))).(((((((((((((((((((.......((((....)))))))))))))))))))))))(((((....)))))............. ( -44.21) >DroSec_CAF1 11284 98 + 1 GAUGAGGACCACACGCAAACACACAGAAUGCCCACACUGCGUAUGAGUAAUCCGACUGGCAAGCAAUUGCGUUACUCAUACGCAGUGUGG---------------CCCCCC---AA ..((.((.......(((..(.....)..)))((((((((((((((((((((.......((((....))))))))))))))))))))))))---------------..)).)---). ( -40.81) >DroEre_CAF1 11825 110 + 1 AAAGAGAGCCACACGCAAAUAACCAGAAUGCUCACACUGCGUAUUAGUAAUCCGACUGGCAAGCAAUUGCGUUGCUCAUACGCAGUGUGGGUCGCCCACGGCCACC---CC---AA .....((((....................))))(((((((((((.((((((.......((((....)))))))))).)))))))))))((((.(((...))).)))---).---.. ( -38.96) >DroYak_CAF1 8472 109 + 1 ---GAGAGCCACACGCAAACAAGCGGAGUACCCACACUGCGUAUGAGUAAUCCGACUGGCAAGCAAUCGCGUUACUCAUACGCAGUGUGGGUCGCCGACGGCCACC-CCCC---AA ---..(.(((....((......))((.(.((((((((((((((((((((((.(((.((.....)).))).))))))))))))))))))))))).))...))))...-....---.. ( -55.30) >consensus AAUGAGAGCCACACGCAAACAAACAGAAUGCCCACACUGCGUAUGAGUAAUCCGACUGGCAAGCAAUUGCGUUACUCAUACGCAGUGUGGGUCGCCGACGGCCACC_CCCC___AA .............................((((((((((((((((((((((.(((.((.....)).))).))))))))))))))))))))))........................ (-39.29 = -39.35 + 0.06)



| Location | 10,005,740 – 10,005,856 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -52.18 |
| Consensus MFE | -36.66 |
| Energy contribution | -37.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10005740 116 - 22224390 UUUACGGGGGGUUGGUCGUCGGCGACCCACACUGCGUAUGAGUAACGCAAUUGCUUGCCAGUCGGAUUACUCAUACGCAGUGUGGGCAUUCUGUGUGUUUGCGUGCGGCUCUCAUU ......((((((((((((....))))(((((((((((((((((((.((....))...((....)).)))))))))))))))))))(((..(.....)..)))...))))))))... ( -54.20) >DroSec_CAF1 11284 98 - 1 UU---GGGGGG---------------CCACACUGCGUAUGAGUAACGCAAUUGCUUGCCAGUCGGAUUACUCAUACGCAGUGUGGGCAUUCUGUGUGUUUGCGUGUGGUCCUCAUC ..---.(((((---------------(((((((((((((((((((.((....))...((....)).)))))))))))))))(..(((((.....)))))..)..)))))))))... ( -50.20) >DroEre_CAF1 11825 110 - 1 UU---GG---GGUGGCCGUGGGCGACCCACACUGCGUAUGAGCAACGCAAUUGCUUGCCAGUCGGAUUACUAAUACGCAGUGUGAGCAUUCUGGUUAUUUGCGUGUGGCUCUCUUU ..---((---(((.((((..((..(((((((((((((((((((((.....)))))).((....)).......))))))))))))........)))..))..)).)).))))).... ( -42.60) >DroYak_CAF1 8472 109 - 1 UU---GGGG-GGUGGCCGUCGGCGACCCACACUGCGUAUGAGUAACGCGAUUGCUUGCCAGUCGGAUUACUCAUACGCAGUGUGGGUACUCCGCUUGUUUGCGUGUGGCUCUC--- ..---.(((-(((.((((((((.((((((((((((((((((((((..((((((.....))))))..))))))))))))))))))))).).))).......))).)).))))))--- ( -61.71) >consensus UU___GGGG_GGUGGCCGUCGGCGACCCACACUGCGUAUGAGUAACGCAAUUGCUUGCCAGUCGGAUUACUCAUACGCAGUGUGGGCAUUCUGUGUGUUUGCGUGUGGCUCUCAUU .........((((.((.....)).))))(((((((((((((((((.((....))...((....)).)))))))))))))))))((((....(((......)))....))))..... (-36.66 = -37.23 + 0.56)



| Location | 10,005,780 – 10,005,896 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.03 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10005780 116 + 22224390 GUAUGAGUAAUCCGACUGGCAAGCAAUUGCGUUACUCAUACGCAGUGUGGGUCGCCGACGACCAACCCCCCGUAAAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCUA (((((((((((.......((((....)))))))))))))))(((...(((((((....)))))........(((((((((((...))))).))))))....))...)))....... ( -35.21) >DroSec_CAF1 11324 98 + 1 GUAUGAGUAAUCCGACUGGCAAGCAAUUGCGUUACUCAUACGCAGUGUGG---------------CCCCCC---AAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCCA (((((((((((.......((((....)))))))))))))))((.((((((---------------(.....---........)))))))......(((((....)))))...)).. ( -29.33) >DroEre_CAF1 11865 110 + 1 GUAUUAGUAAUCCGACUGGCAAGCAAUUGCGUUGCUCAUACGCAGUGUGGGUCGCCCACGGCCACC---CC---AAAUGUAUGCUAUAGAUUUUCGCAUGUCAAUAUGCUUAGCCA .................(((((((((((((((.((.((((((...((.((((.(((...))).)))---))---)..)))))).....(.....)))))).)))).))))).))). ( -32.10) >DroYak_CAF1 8509 112 + 1 GUAUGAGUAAUCCGACUGGCAAGCAAUCGCGUUACUCAUACGCAGUGUGGGUCGCCGACGGCCACC-CCCC---AAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCUA (((((((((((.(((.((.....)).))).)))))))))))((.(.(.((((.(((...))).)))-).))---((((((((...))))))))..(((((....)))))...)).. ( -36.80) >consensus GUAUGAGUAAUCCGACUGGCAAGCAAUUGCGUUACUCAUACGCAGUGUGGGUCGCCGACGGCCACC_CCCC___AAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCCA (((((((((((.(((.((.....)).))).)))))))))))(((((((((((((....)))))..............(((((((...........))))).)).))))))..)).. (-27.80 = -28.05 + 0.25)


| Location | 10,005,816 – 10,005,925 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -16.60 |
| Energy contribution | -17.91 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10005816 109 + 22224390 CAUACGCAGUGUGGGUCGCCGACGACCAACCCCCCGUAAAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCUAUCCGCCCCACU----------GCUGUGAUUUCU-CAUCCA ((((.((((((.(((.((...................(((((((((...))))))))).(((((....))))).........)))))))))----------))))))......-...... ( -27.60) >DroSec_CAF1 11360 92 + 1 CAUACGCAGUGUGG---------------CCCCCC---AAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCCAUCCGCCCUACU----------GCUGUGACUUCUCCAUCCA ((((.((((((.((---------------(.....---((((((((...))))))))..(((((....)))))..........))).))))----------))))))............. ( -23.60) >DroEre_CAF1 11901 113 + 1 CAUACGCAGUGUGGGUCGCCCACGGCCACC---CC---AAAUGUAUGCUAUAGAUUUUCGCAUGUCAAUAUGCUUAGCCAUCCGACCCACUGCAGUGCACCGCUGUGAUUCCU-CAUCCA ....((((((((((((((.....(((....---..---((((.(((...))).))))..(((((....)))))...)))...))))))))(((...)))..))))))......-...... ( -30.10) >DroYak_CAF1 8545 105 + 1 CAUACGCAGUGUGGGUCGCCGACGGCCACC-CCCC---AAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCUAUCCACCCAACU----------GCUGUGAUUCCU-CAUCCA ((((.(((((.(((((....((..((....-....---((((((((...))))))))..(((((....)))))...))..)).))))))))----------))))))......-...... ( -27.60) >consensus CAUACGCAGUGUGGGUCGCCGACGGCCACC_CCCC___AAAUGUAUGCUAUACAUUUUUGCAUGUCAAUAUGCUUAGCCAUCCGCCCCACU__________GCUGUGAUUCCU_CAUCCA ....((((((((((((.((...................((((((((...))))))))..(((((....)))))...)).))))))................))))))............. (-16.60 = -17.91 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:05 2006