| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,975,917 – 9,976,007 |
| Length | 90 |
| Max. P | 0.844310 |

| Location | 9,975,917 – 9,976,007 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.30 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -10.81 |
| Energy contribution | -12.71 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
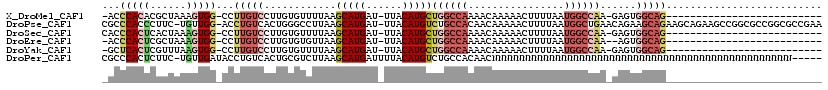
>X_DroMel_CAF1 9975917 90 + 22224390 -ACCCACACGCUAAAGUGG-CCUUGUCCUUGUGUUUUAAGCAUGAU-UUACAUGCUGGCCAAAACAAAAACUUUUAAUGGCCAA-GAGUGGCAG-------------------------- -........((((...(((-(((((......((((((.((((((..-...))))))....))))))........))).))))).-...))))..-------------------------- ( -25.14) >DroPse_CAF1 32704 117 + 1 CGCCCACCCUUC-UGUUGG-ACCUGUCACUGGGCCUUAAGCAUGAU-UUACAUGUCUGCCACAACAAAAACUUUUAAUGGCUGAACAGAAGCAGAAGCAGAAGCCGGCGCCGGCGCCGAA ........((((-((((..-..((((..(((..(.....(((((..-...)))))..((((................)))).)..)))..)))).)))))))).(((((....))))).. ( -33.79) >DroSec_CAF1 21260 91 + 1 CACCCACUCACUAAAGUGG-CCUUGUCCUUGUGUUUUAAGCAUGAU-UUACAUGCUGGCCAAAACAAAAACUUUUAAUGGCCAA-GAGUGGCAG-------------------------- ...((((((((....))((-(((((......((((((.((((((..-...))))))....))))))........))).))))..-))))))...-------------------------- ( -27.04) >DroEre_CAF1 8584 89 + 1 -ACCCACUCGCUAAAGUGG-CCUUGUCCUUGUGUGUUAAGCAUGAU-UUACAUGCUGGCCAAAACAAAAACUUUUAAUGGCCAA--AGUGGCAG-------------------------- -........((((...(((-((..((..((((..((((.(((((..-...)))))))))....))))..)).......))))).--..))))..-------------------------- ( -25.00) >DroYak_CAF1 8876 90 + 1 -GCUCACUCGUUUAAGUGG-CCUUGUCCUUGUGUUUUAAGCAUGAU-UUACAUGCUGGCCAAAACAAAAACUUUUAAUGGCCAA-GAGUGGCAG-------------------------- -((.(((((.......(((-(((((......((((((.((((((..-...))))))....))))))........))).))))).-)))))))..-------------------------- ( -28.14) >DroPer_CAF1 33435 114 + 1 CGCCCACUCUUC-UGUUGAUACCUGUCACUGCGUCUUAAGCAUGAUUUUACAUGUCUGCCACAACNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN----- ............-.((.((((...((((.(((.......)))))))......)))).))........................................................----- ( -7.40) >consensus _ACCCACUCGCUAAAGUGG_CCUUGUCCUUGUGUUUUAAGCAUGAU_UUACAUGCUGGCCAAAACAAAAACUUUUAAUGGCCAA_GAGUGGCAG__________________________ ...(((((......)))))...(((((............(((((......)))))((((((................))))))......))))).......................... (-10.81 = -12.71 + 1.89)


| Location | 9,975,917 – 9,976,007 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.30 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -12.97 |
| Energy contribution | -15.20 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9975917 90 - 22224390 --------------------------CUGCCACUC-UUGGCCAUUAAAAGUUUUUGUUUUGGCCAGCAUGUAA-AUCAUGCUUAAAACACAAGGACAAGG-CCACUUUAGCGUGUGGGU- --------------------------...((((.(-((((((.((....((((((((.((....((((((...-..))))))...)).))))))))))))-)))....))...))))..- ( -24.50) >DroPse_CAF1 32704 117 - 1 UUCGGCGCCGGCGCCGGCUUCUGCUUCUGCUUCUGUUCAGCCAUUAAAAGUUUUUGUUGUGGCAGACAUGUAA-AUCAUGCUUAAGGCCCAGUGACAGGU-CCAACA-GAAGGGUGGGCG .((((((....)))))).....(((..(.((((((((..(((((.....))..(..(((.(((((.((((...-..))))))....))))))..)..)))-..))))-)))).)..))). ( -37.70) >DroSec_CAF1 21260 91 - 1 --------------------------CUGCCACUC-UUGGCCAUUAAAAGUUUUUGUUUUGGCCAGCAUGUAA-AUCAUGCUUAAAACACAAGGACAAGG-CCACUUUAGUGAGUGGGUG --------------------------...((((((-..((((.((....((((((((.((....((((((...-..))))))...)).))))))))))))-))((....))))))))... ( -28.50) >DroEre_CAF1 8584 89 - 1 --------------------------CUGCCACU--UUGGCCAUUAAAAGUUUUUGUUUUGGCCAGCAUGUAA-AUCAUGCUUAACACACAAGGACAAGG-CCACUUUAGCGAGUGGGU- --------------------------...(((((--((((((.((....((((((((..((...((((((...-..))))))...)).))))))))))))-))).......))))))..- ( -27.41) >DroYak_CAF1 8876 90 - 1 --------------------------CUGCCACUC-UUGGCCAUUAAAAGUUUUUGUUUUGGCCAGCAUGUAA-AUCAUGCUUAAAACACAAGGACAAGG-CCACUUAAACGAGUGAGC- --------------------------..(((((((-.(((((.((....((((((((.((....((((((...-..))))))...)).))))))))))))-))).......))))).))- ( -25.60) >DroPer_CAF1 33435 114 - 1 -----NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUUGUGGCAGACAUGUAAAAUCAUGCUUAAGACGCAGUGACAGGUAUCAACA-GAAGAGUGGGCG -----..................................................((..(.((((.((((......))))))......)).)..))..((....((.-.....))..)). ( -7.20) >consensus __________________________CUGCCACUC_UUGGCCAUUAAAAGUUUUUGUUUUGGCCAGCAUGUAA_AUCAUGCUUAAAACACAAGGACAAGG_CCACUUUAACGAGUGGGC_ ............................((((.....))))........((((((((.((....((((((......))))))...)).)))))))).....(((((......)))))... (-12.97 = -15.20 + 2.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:54 2006