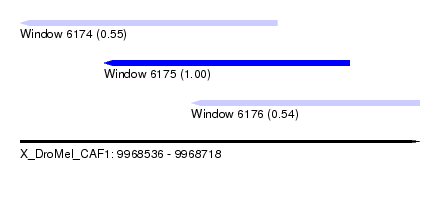
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,968,536 – 9,968,718 |
| Length | 182 |
| Max. P | 0.998577 |

| Location | 9,968,536 – 9,968,653 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -19.75 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9968536 117 - 22224390 ACCAACUAUCCA-ACCCACUUUUCUGUGGGAAAAUCCCGAACGAAACUCAACAAAGAGGUUUUUUUUUUCGUUUUUGGCCAAGGCUUUUUCGCACACCUGUCCAGGUGGUAAAUUUGU-- .((((.......-.(((((......)))))........(((((((((((......)))........)))))))))))).((((((......)).(((((....))))).....)))).-- ( -26.90) >DroSec_CAF1 9013 111 - 1 ACCGACUAUCCA-ACCCACUUUUCUGUGGGAAAAUCCCGAACGAAACUCAACAAAGAGGUUUUUU--------UUUGGCCAAGGCUUUUUCGCACACCUGUCCAGAUGGUAAAUUUUUUA ....((((((..-.(((((......)))))...........((((((((......))).......--------...(((....))).)))))............)))))).......... ( -23.00) >DroEre_CAF1 1180 116 - 1 ACCGACUGUGCA-ACCCACUUUGCUGUGGGAAAAACC-CAACGAAACUCCACAAAGAAAAGGUUUUCUUAUUUUUUGGCCAAGGCUUUUUCACACACCUGUCCAGGUGGUAAAUUUGU-- ((((.(((..((-.(((((......))))).......-....((((.....(((((((.(((....))).)))))))((....))..)))).......))..))).))))........-- ( -28.20) >DroYak_CAF1 1184 114 - 1 ACCGACGGUCCAAACCCACUUUUCUGUGGGAAAAACC-AAACGGAAUUCAACAAAGAAAAUGUUUUUU---UUUCUUGCCAAGGCUUUUUCGCACACCUGUACAGGUGGUAAAUUGGU-- .(((..(((.....(((((......)))))....)))-...))).........(((((((.......)---))))))(((((.((......)).(((((....))))).....)))))-- ( -30.90) >consensus ACCGACUAUCCA_ACCCACUUUUCUGUGGGAAAAACC_GAACGAAACUCAACAAAGAAAAUGUUUUUU___UUUUUGGCCAAGGCUUUUUCGCACACCUGUCCAGGUGGUAAAUUUGU__ ..............(((((......)))))............(((((((......)))...................((....))..))))((.(((((....))))))).......... (-19.75 = -19.62 + -0.12)


| Location | 9,968,574 – 9,968,686 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.57 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -10.26 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9968574 112 - 22224390 UUUCUCAUAACCCUUCCCC-------CAAAAAACCAAAAAACCAACUAUCCA-ACCCACUUUUCUGUGGGAAAAUCCCGAACGAAACUCAACAAAGAGGUUUUUUUUUUCGUUUUUGGCC ..................(-------((((((...((((((((.........-.(((((......)))))........((.......))........))))))))......))))))).. ( -20.90) >DroSec_CAF1 9053 111 - 1 UUUCUCAUAACCCUUCCCAACAACAAAAAAAAACCAAAAAACCGACUAUCCA-ACCCACUUUUCUGUGGGAAAAUCCCGAACGAAACUCAACAAAGAGGUUUUUU--------UUUGGCC .......................((((((((((((.................-.(((((......)))))........((.......))........))))))))--------))))... ( -22.00) >DroEre_CAF1 1218 100 - 1 -UUCUCACAACACUUCUCC-------CA----------AAACCGACUGUGCA-ACCCACUUUGCUGUGGGAAAAACC-CAACGAAACUCCACAAAGAAAAGGUUUUCUUAUUUUUUGGCC -.................(-------((----------(((.....((((..-.(((((......))))).......-....(.....)))))((((((....))))))...)))))).. ( -16.40) >DroYak_CAF1 1222 98 - 1 -UUCUCAUAACACUUCGCC-------CA----------AAACCGACGGUCCAAACCCACUUUUCUGUGGGAAAAACC-AAACGGAAUUCAACAAAGAAAAUGUUUUUU---UUUCUUGCC -...............((.-------..----------...(((..(((.....(((((......)))))....)))-...))).........(((((((.......)---)))))))). ( -18.90) >consensus _UUCUCAUAACACUUCCCC_______CA__________AAACCGACUAUCCA_ACCCACUUUUCUGUGGGAAAAACC_GAACGAAACUCAACAAAGAAAAUGUUUUUU___UUUUUGGCC .........................................((((.........(((((......)))))................(((......)))................)))).. (-10.26 = -10.57 + 0.31)


| Location | 9,968,614 – 9,968,718 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9968614 104 - 22224390 GUUUCUUGGUUUGCAUGGCAUGCUGCACAAUUUUUCUCAUAACCCUUCCCC-------CAAAAAACCAAAAAACCAACUAUCCA-ACCCACUUUUCUGUGGGAAAAUCCCGA ((((.(((((((...(((................................)-------))..))))))).))))..........-.(((((......))))).......... ( -18.25) >DroSec_CAF1 9085 106 - 1 GUUUCUGGGUUUGC-----AUGCUGCACAAUUUUUCUCAUAACCCUUCCCAACAACAAAAAAAAACCAAAAAACCGACUAUCCA-ACCCACUUUUCUGUGGGAAAAUCCCGA ...((.((((((((-----.....))..........................................................-.(((((......))))).)))))).)) ( -16.00) >DroEre_CAF1 1258 87 - 1 GUUUCUAGGCUUGC-----AUGCUGCAGAAUU-UUCUCACAACACUUCUCC-------CA----------AAACCGACUGUGCA-ACCCACUUUGCUGUGGGAAAAACC-CA ..((((.(((....-----..)))..))))((-(((((((((((..((...-------..----------.....)).)))(((-(......))))))))))))))...-.. ( -18.00) >DroYak_CAF1 1259 88 - 1 GUUUCUGGGCUUGC-----AUGCUGCACAAUU-UUCUCAUAACACUUCGCC-------CA----------AAACCGACGGUCCAAACCCACUUUUCUGUGGGAAAAACC-AA ((((.(((((.(((-----.....)))....(-(......))......)))-------))----------))))....(((.....(((((......)))))....)))-.. ( -20.70) >consensus GUUUCUGGGCUUGC_____AUGCUGCACAAUU_UUCUCAUAACACUUCCCC_______CA__________AAACCGACUAUCCA_ACCCACUUUUCUGUGGGAAAAACC_GA ........((..((.......)).))............................................................(((((......))))).......... (-10.35 = -10.10 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:44 2006