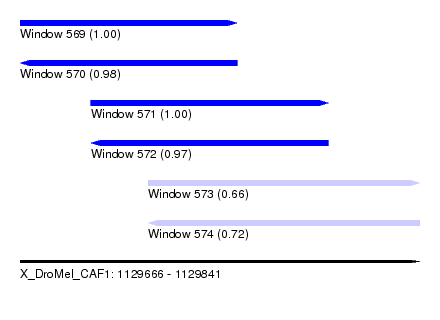
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,129,666 – 1,129,841 |
| Length | 175 |
| Max. P | 0.996344 |

| Location | 1,129,666 – 1,129,761 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1129666 95 + 22224390 UUCCAGCUUCAGUCCGGGGUUCAUUAAUGCGCACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUC-UUUUGU ...(((((.(((((.((......((((.((((.....))))))))((((((.....))))))..))))))).......)))))......-...... ( -26.90) >DroSec_CAF1 26288 93 + 1 UUCCAGCUUCAGUC-GGGGUUCAUUAAUGCGCACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGCCUGUCUUUCCAGCUGUCUCU--UUUUGU ...(((((..((.(-(((.....((((.((((.....))))))))((((((.....)))))).....)))).))....))))).....--...... ( -26.70) >DroSim_CAF1 27736 93 + 1 UUCCAGCUUCAGUC-GGGGUUCAUUAAUGCGCACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGCCUGUCUUUCCAGCUGUCUCU--UUUUGU ...(((((..((.(-(((.....((((.((((.....))))))))((((((.....)))))).....)))).))....))))).....--...... ( -26.70) >DroEre_CAF1 27601 95 + 1 UCCCAGCUUCAGUC-GGUGUUCAUUAAUGCGCACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUCUUUUUGU ...(((((.(((((-(((.....((((.((((.....))))))))((((((.....)))))).)))))))).......)))))............. ( -32.20) >DroYak_CAF1 27196 95 + 1 UGCCAGCUUCAGUC-GGGGUUCAUUAAUGCGCACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUCUUUUUGU ...(((((.(((((-((......((((.((((.....))))))))((((((.....))))))..))))))).......)))))............. ( -30.70) >consensus UUCCAGCUUCAGUC_GGGGUUCAUUAAUGCGCACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUC_UUUUGU ...(((((.(((((.((......((((.((((.....))))))))((((((.....))))))..))))))).......)))))............. (-24.52 = -24.92 + 0.40)

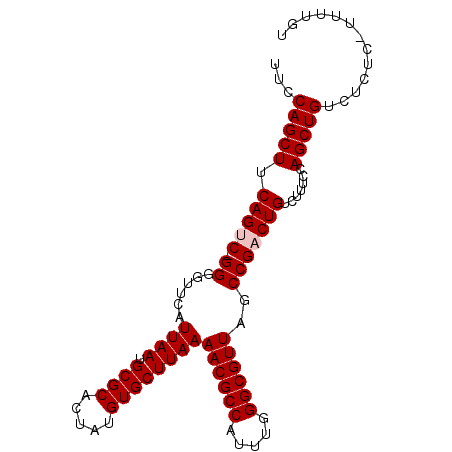
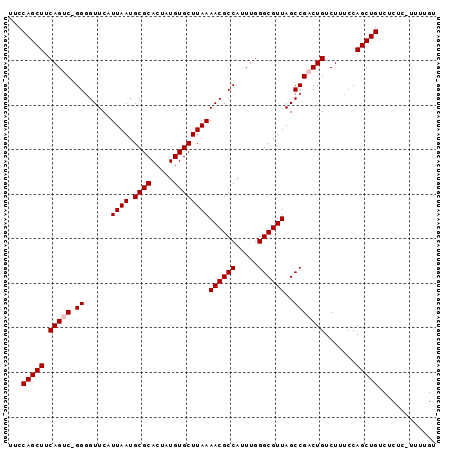
| Location | 1,129,666 – 1,129,761 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1129666 95 - 22224390 ACAAAA-GAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUGCGCAUUAAUGAACCCCGGACUGAAGCUGGAA ......-......(((((.......((((((((....)))....(((.((((...((((...))))........)))).)))))))).)))))... ( -25.30) >DroSec_CAF1 26288 93 - 1 ACAAAA--AGAGACAGCUGGAAAGACAGGCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUGCGCAUUAAUGAACCCC-GACUGAAGCUGGAA ......--.....(((((....((...((.((..((((((.....))))))....((((...))))...........))))-..))..)))))... ( -23.70) >DroSim_CAF1 27736 93 - 1 ACAAAA--AGAGACAGCUGGAAAGACAGGCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUGCGCAUUAAUGAACCCC-GACUGAAGCUGGAA ......--.....(((((....((...((.((..((((((.....))))))....((((...))))...........))))-..))..)))))... ( -23.70) >DroEre_CAF1 27601 95 - 1 ACAAAAAGAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUGCGCAUUAAUGAACACC-GACUGAAGCUGGGA .............(((((.......(((((((..((((((.....))))))....((((...)))).............))-))))).)))))... ( -28.50) >DroYak_CAF1 27196 95 - 1 ACAAAAAGAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUGCGCAUUAAUGAACCCC-GACUGAAGCUGGCA ...............((..(.....(((((((..((((((.....))))))....((((...)))).............))-)))))...)..)). ( -29.00) >consensus ACAAAA_GAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUGCGCAUUAAUGAACCCC_GACUGAAGCUGGAA .............(((((.......(((((((..((((((.....))))))....((((...)))).............)).))))).)))))... (-22.30 = -22.70 + 0.40)

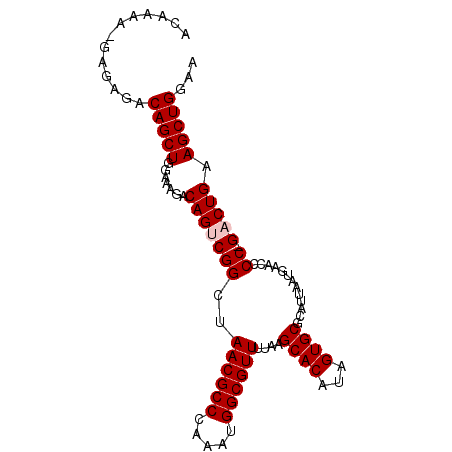
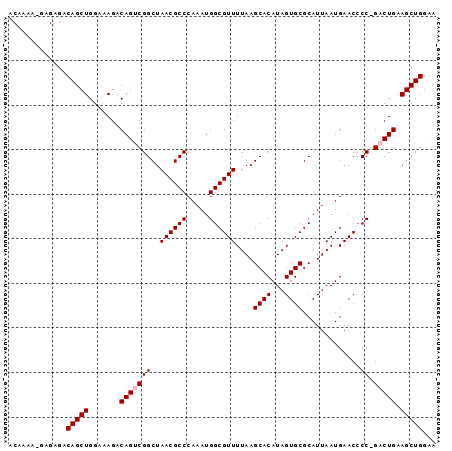
| Location | 1,129,697 – 1,129,801 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1129697 104 + 22224390 CACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUC-UUUUGUGAACAGAAGUGGAUCUCAGACACUCGGAAAAAUGGCUCAA .....((.(((...((((((.....))))))..((((.(((((.((((.((((.((.(-....).))))))...))))....))))).))))......))).)). ( -32.80) >DroSec_CAF1 26318 103 + 1 CACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGCCUGUCUUUCCAGCUGUCUCU--UUUUGUGAGCAGAAGUGGAUCUCAGACACUCGCAAAAAUGGCUCAA .................(((((((.(((....)))((.(((((.((((.(((.(((.--......))))))...))))....)))))...))..))))))).... ( -30.80) >DroSim_CAF1 27766 103 + 1 CACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGCCUGUCUUUCCAGCUGUCUCU--UUUUGUGAGCAGAAGUGGAUCUCAGACACUCGCAAAAAUGGCUCAA .................(((((((.(((....)))((.(((((.((((.(((.(((.--......))))))...))))....)))))...))..))))))).... ( -30.80) >DroEre_CAF1 27631 105 + 1 CACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUCUUUUUGUGAGCAGAAGUGGAUCUCAGACACUUGGAAAAAUGGUCCAA ......(.(((...((((((.....))))))))))((((((.(((((((.(((((((.((((((....)))))).))).....)))).))))))).))))))... ( -33.30) >DroYak_CAF1 27226 105 + 1 CACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUCUUUUUGUGAGCAGCCGUGGAUCUCAGACACUCGGAAAAAUGGCUCAA .....((.(((...((((((.....))))))..((((.(((((.((((((((.(((.(.....).)))))))..))))....))))).))))......))).)). ( -37.60) >consensus CACUAUGUGCUUAAAACGCCAUUUGGGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUC_UUUUGUGAGCAGAAGUGGAUCUCAGACACUCGGAAAAAUGGCUCAA .....((.(((...((((((.....))))))..((((.(((((.((((.(((.(((.........))))))...))))....))))).))))......))).)). (-28.22 = -28.90 + 0.68)

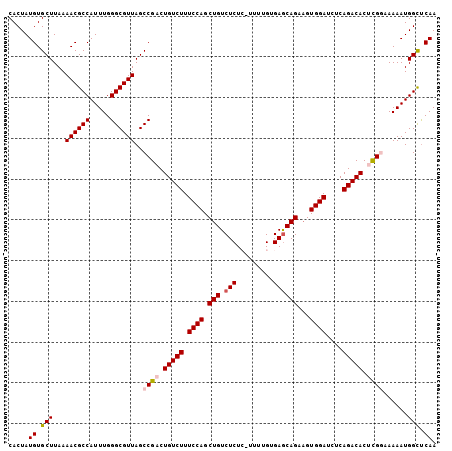
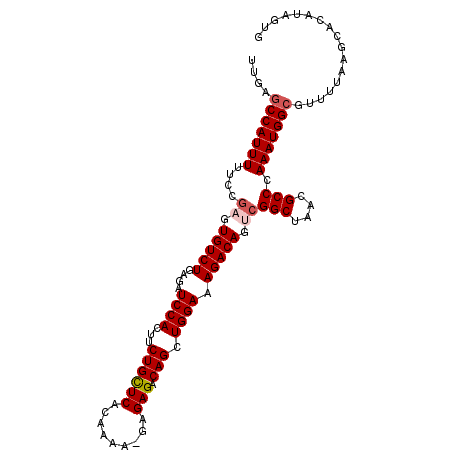
| Location | 1,129,697 – 1,129,801 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -27.24 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1129697 104 - 22224390 UUGAGCCAUUUUUCCGAGUGUCUGAGAUCCACUUCUGUUCACAAAA-GAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUG ....(((((((....((.(((((....(((((((((.(((......-)))))).)).)))).))))).))(((....))).)))))))................. ( -31.00) >DroSec_CAF1 26318 103 - 1 UUGAGCCAUUUUUGCGAGUGUCUGAGAUCCACUUCUGCUCACAAAA--AGAGACAGCUGGAAAGACAGGCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUG ....(((((((....(..(((((....((((...((((((......--.))).))).)))).)))))..)(((....))).)))))))................. ( -30.00) >DroSim_CAF1 27766 103 - 1 UUGAGCCAUUUUUGCGAGUGUCUGAGAUCCACUUCUGCUCACAAAA--AGAGACAGCUGGAAAGACAGGCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUG ....(((((((....(..(((((....((((...((((((......--.))).))).)))).)))))..)(((....))).)))))))................. ( -30.00) >DroEre_CAF1 27631 105 - 1 UUGGACCAUUUUUCCAAGUGUCUGAGAUCCACUUCUGCUCACAAAAAGAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUG ...(((..((((((((..(((((.(((......))).(((.......))))))))..))))))))..))).(((((((((.....))))))...)))........ ( -31.60) >DroYak_CAF1 27226 105 - 1 UUGAGCCAUUUUUCCGAGUGUCUGAGAUCCACGGCUGCUCACAAAAAGAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUG ....(((((((....((.(((((....((((..(((((((.(.....).))).)))))))).))))).))(((....))).)))))))................. ( -36.20) >consensus UUGAGCCAUUUUUCCGAGUGUCUGAGAUCCACUUCUGCUCACAAAA_GAGAGACAGCUGGAAAGACAGUCGGCUAACGCCCAAAUGGCGUUUUAAGCACAUAGUG ....(((((((....((.(((((....((((...((((((.........))).))).)))).))))).))(((....))).)))))))................. (-27.24 = -27.88 + 0.64)

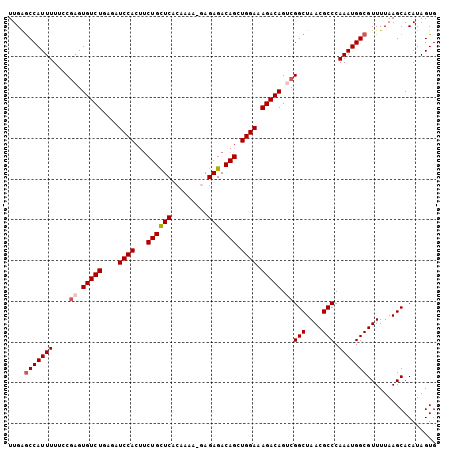
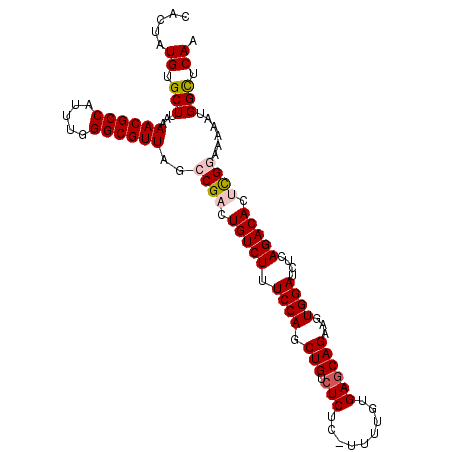
| Location | 1,129,722 – 1,129,841 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.92 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1129722 119 + 22224390 GGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUC-UUUUGUGAACAGAAGUGGAUCUCAGACACUCGGAAAAAUGGCUCAAGGGCCAGAAGAAAACUGUGCAUUCUUUUAGCUCUAAAUGU (((....)))((((((.(((((((.((((((((-(((((....)))))).))).....)))))).))))).)))).)).(((((.(((((((.........))))))).)))))...... ( -35.40) >DroSec_CAF1 26343 117 + 1 GGCGUUAGCCGCCUGUCUUUCCAGCUGUCUCU--UUUUGUGAGCAGAAGUGGAUCUCAGACACUCGCAAAAAUGGCUCAAGUGCCAGAAGAACACUUUU-GCUUUUUAAGCUCUAAAUAA (((....)))((.(((((.((((.(((.(((.--......))))))...))))....)))))...((((((.((((......)))).........))))-)).......))......... ( -30.40) >DroSim_CAF1 27791 117 + 1 GGCGUUAGCCGCCUGUCUUUCCAGCUGUCUCU--UUUUGUGAGCAGAAGUGGAUCUCAGACACUCGCAAAAAUGGCUCAAGUGCCAGAAGAAAACUUUU-GCUUUUUAAGCUCUAAAUAA (((....)))((.(((((.((((.(((.(((.--......))))))...))))....)))))...((((((.((((......)))).........))))-)).......))......... ( -30.40) >DroEre_CAF1 27656 109 + 1 GGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUCUUUUUGUGAGCAGAAGUGGAUCUCAGACACUUGGAAAAAUGGUCCAAGUGCCAGUGGGAAAAA-----------UAAAACUUAAUAC (((....)))...(((.(((((.((((..(((.((((((....)))))).))).......((((((((.......)))))))).)))).))))).)-----------))........... ( -31.00) >DroYak_CAF1 27251 95 + 1 GGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUCUUUUUGUGAGCAGCCGUGGAUCUCAGACACUCGGAAAAAUGGCUCAAGUGCCAGAUG-------------------------AAAAC ..((((..((((.(((((.((((((((.(((.(.....).)))))))..))))....))))).)))).....((((......))))))))-------------------------..... ( -32.70) >consensus GGCGUUAGCCGACUGUCUUUCCAGCUGUCUCUC_UUUUGUGAGCAGAAGUGGAUCUCAGACACUCGGAAAAAUGGCUCAAGUGCCAGAAGAAAACU_U____U_UUUAAGCUCUAAAUAA (((....)))((.(((((.((((.(((.(((.........))))))...))))....))))).)).......((((......)))).................................. (-24.20 = -24.48 + 0.28)

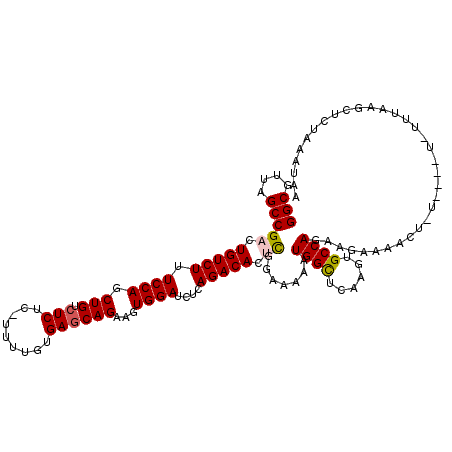
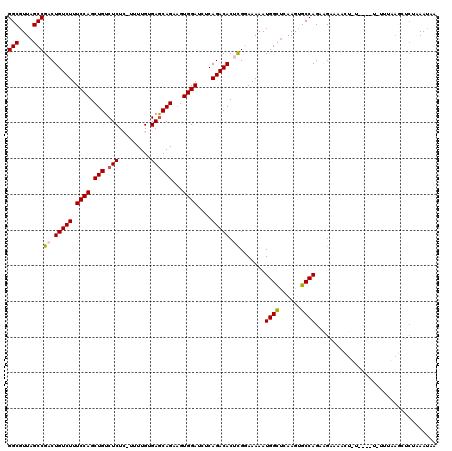
| Location | 1,129,722 – 1,129,841 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.92 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.36 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1129722 119 - 22224390 ACAUUUAGAGCUAAAAGAAUGCACAGUUUUCUUCUGGCCCUUGAGCCAUUUUUCCGAGUGUCUGAGAUCCACUUCUGUUCACAAAA-GAGAGACAGCUGGAAAGACAGUCGGCUAACGCC ...((.((.((((.(((((.((...)).))))).)))).)).))...........((.(((((....(((((((((.(((......-)))))).)).)))).))))).))(((....))) ( -29.50) >DroSec_CAF1 26343 117 - 1 UUAUUUAGAGCUUAAAAAGC-AAAAGUGUUCUUCUGGCACUUGAGCCAUUUUUGCGAGUGUCUGAGAUCCACUUCUGCUCACAAAA--AGAGACAGCUGGAAAGACAGGCGGCUAACGCC .........((((.....((-((((((((((...........))).)))))))))....((((....((((...((((((......--.))).))).)))).))))))))(((....))) ( -33.10) >DroSim_CAF1 27791 117 - 1 UUAUUUAGAGCUUAAAAAGC-AAAAGUUUUCUUCUGGCACUUGAGCCAUUUUUGCGAGUGUCUGAGAUCCACUUCUGCUCACAAAA--AGAGACAGCUGGAAAGACAGGCGGCUAACGCC .........((((.....((-(((((........((((......)))))))))))....((((....((((...((((((......--.))).))).)))).))))))))(((....))) ( -31.70) >DroEre_CAF1 27656 109 - 1 GUAUUAAGUUUUA-----------UUUUUCCCACUGGCACUUGGACCAUUUUUCCAAGUGUCUGAGAUCCACUUCUGCUCACAAAAAGAGAGACAGCUGGAAAGACAGUCGGCUAACGCC .......(((((.-----------..((((.....((((((((((.......)))))))))).))))(((((((((.(((.......)))))).)).)))))))))....(((....))) ( -31.50) >DroYak_CAF1 27251 95 - 1 GUUUU-------------------------CAUCUGGCACUUGAGCCAUUUUUCCGAGUGUCUGAGAUCCACGGCUGCUCACAAAAAGAGAGACAGCUGGAAAGACAGUCGGCUAACGCC .....-------------------------....((((......)))).......((.(((((....((((..(((((((.(.....).))).)))))))).))))).))(((....))) ( -31.60) >consensus GUAUUUAGAGCUAAAA_A____A_AGUUUUCUUCUGGCACUUGAGCCAUUUUUCCGAGUGUCUGAGAUCCACUUCUGCUCACAAAA_GAGAGACAGCUGGAAAGACAGUCGGCUAACGCC ..................................((((......)))).......((.(((((....((((...((((((.........))).))).)))).))))).))(((....))) (-22.72 = -23.36 + 0.64)

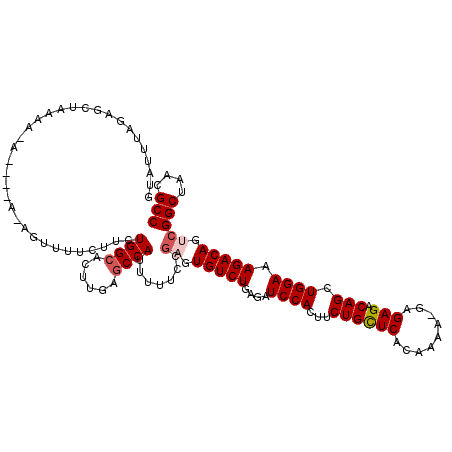
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:03 2006